ajax不利于seo
Lately, my parents will often bring up in conversation their desire to move away from their California home and find a new place to settle down for retirement. Typically they will cite factors that they perceive as having altered the essence of their suburban and rural county such as higher costs of living, population growth, worsening traffic, and growing rates of violent crime that they say have made them feel estranged from their community in recent years. Despite these concerns, both admit that there is much they would miss about their current home such as the mild climate of the California coast, the abundant natural beauty of the region, and an vast matrix of open space and public lands that allow them to enjoy outdoor hobbies throughout the year.
最近,我的父母經常會談論他們想離開加利福尼亞州的家,并找到一個定居下來退休的新地方的愿望。 通常,他們會引用自己認為改變了郊區和鄉村縣城本質的因素,例如更高的生活成本,人口增長,交通狀況惡化以及暴力犯罪率上升,他們說這使他們對自己的社區感到疏遠。最近幾年。 盡管存在這些擔憂,但他們倆都承認,他們目前的住所會遺漏很多東西,例如加利福尼亞海岸的溫和氣候,該地區豐富的自然風光以及廣闊的開放空間和公共土地,使他們能夠享受全年都有戶外活動。
The idea for Niche came from wanting to help my parents explore their relocation options by taking into consideration what matters most to them about the place they live now. The name of this app is taken from a foundational concept in biology. While many definitions of a niche exist, from verbal models to mathematical frameworks, virtually all definitions agree that one’s niche can be thought of as the set of environmental conditions necessary for an organism to thrive. While some species can only tolerate a narrow range of conditions (think temperature, light, salinity, etc.) others will do just fine when exposed to large changes in these same variables. But even members of the same species will occupy a different niche due to the individual variation that exists in their anatomy, physiology, and behavior. The point is that no two individuals experience a shared environment in exactly the same way. We all have different tolerances.
Niche的想法來自希望通過考慮父母對他們現在住的地方最重要的問題來幫助我的父母探索他們的搬遷選擇。 該應用程序的名稱來自生物學的基本概念。 盡管存在許多關于生態位的定義,從語言模型到數學框架,但幾乎所有定義都同意,人們的生態位可以被認為是有機體賴以生存的一系列環境條件。 雖然某些物種只能忍受狹窄的條件(例如溫度,光照,鹽度等),但其他物種在暴露于這些相同變量的較大變化時也可以正常工作。 但是,即使是同一物種的成員,由于其解剖結構,生理和行為方面的個體差異,它們也會占據不同的位置。 關鍵是,沒有兩個人會以完全相同的方式體驗共享的環境。 我們都有不同的容忍度。
Borrowing from this concept, it is easy to imagine how our own ability to thrive will be influenced by our surroundings. Niche works by asking users to identify a location where they would like to live and answer a few questions about what is important to them about that place. The program then identifies other areas in the country with similar features before making recommendations on relocation options. Niche considers both the more salient characteristics of a location such as the climate and land uses as well as factors that may be less conspicuous such as economic, demographic, and political indicators. Niche relies on the belief that the best tools to guide our understanding of the conditions best suit us as individuals will come from reflecting on our experiences. You know what you want to find in the places you want to live and Niche can help you find them.
從這個概念中借用,很容易想象我們的生存能力將受到周圍環境的影響。 Niche的工作原理是要求用戶確定他們想要居住的位置,并回答一些有關該位置對他們而言重要的問題。 然后,該程序在建議搬遷選項之前,先確定該國其他具有類似特征的地區。 Niche既考慮了地理位置的突出特征,例如氣候和土地利用,也考慮了經濟,人口和政治指標等不太明顯的因素。 Niche依靠這樣一種信念,即指導個人對條件的理解的最佳工具最適合我們,因為個人會從反思我們的經驗中獲得。 您知道要在自己想住的地方找到什么,Niche可以幫助您找到它們。
All of the data preparation and analysis was performed using R because I knew that I would create the interactive front-end with Shiny. Below, I detail the data and methods that went into creating Niche, but do not cover what went into creating the Shiny app. If you would like to know more about that, you can find that code as well as the rest of the project source code on my GitHub repository. If you have any feedback, questions, or suggestions please get in touch with me by email.
所有數據準備和分析都是使用R進行的,因為我知道我將使用Shiny創建交互式前端。 下面,我詳細介紹創建Niche所需的數據和方法,但不介紹創建Shiny應用程序所需的內容。 如果您想了解更多信息,可以在我的GitHub存儲庫中找到該代碼以及項目源代碼的其余部分。 如果您有任何反饋,問題或建議,請通過電子郵件與我聯系 。
0.獲取縣級邊界數據,此分析的單位 (0. Get County-level Boundary Data, the unit of this analysis)
Before getting started, I needed to aquire a geospatial dataset for all of the counties in the contiguous 48 U.S. states. Fortunately, ggplot2 packages makes this easy with the ggplot2::map_data() function. However, I knew that I would eventually like to have the data as an ‘sf’ type object used by the sf (“simple features”) geospatial package. Therefore, I had to first download the county level data using ggplot2, convert the coordinate data into a ’Spatial*’ object (used by the sp package), save this data to local storage as a shapefile, and finally read the file back into R using sf::st_read(). I decided to save the data as a shapefile as an intermediate data prodcut because the conversion to ‘SpatialPolygonsDataFrame’ was non-trivial (this code helped) and took a bit of time to finish.
開始之前,我需要獲取美國48個連續州的所有縣的地理空間數據集。 幸運的是, ggplot2軟件包通過ggplot2::map_data()函數使此操作變得容易。 但是,我知道我最終希望將數據作為sf (“簡單要素”)地理空間包使用的“ sf”類型對象。 因此,我必須首先使用ggplot2下載縣級數據,將坐標數據轉換為“ Spatial *”對象(由sp程序包使用),然后將此數據作為shapefile保存到本地存儲中,最后將文件讀回到R使用sf::st_read() 。 我決定將數據另存為shapefile作為中間數據產品,因為到“ SpatialPolygonsDataFrame”的轉換非常簡單( 此代碼有所幫助 ),并且花費了一些時間才能完成。
library(ggplot2)
county_df <- map_data("county") %>%
mutate(region = tolower(region)) %>%
mutate(subregion = tolower(subregion)) %>%
mutate(subregion = str_replace(subregion, "st ", "saint ")) %>%
mutate(subregion = str_replace(subregion, "ste ", "saint ")) %>%
mutate(region = str_replace(region, "\\s", "_")) %>%
mutate(subregion = str_replace(subregion, "\\s", "_")) %>%
mutate(id = paste(subregion, region, sep=",_"))
coordinates(county_df) = c("long","lat") #converts to SpatialPointsDataFrame
#convert to SpatialPolygonsDataFrame
#https://stackoverflow.com/questions/21759134/convert-spatialpointsdataframe-to-spatialpolygons-in-r
points2polygons <- function(df,data) {
get.grpPoly <- function(group,ID,df) {
Polygon(coordinates(df[df$id==ID & df$group==group,]))
}
get.spPoly <- function(ID,df) {
Polygons(lapply(unique(df[df$id==ID,]$group),get.grpPoly,ID,df),ID)
}
spPolygons <- SpatialPolygons(lapply(unique(df$id),get.spPoly,df))
SpatialPolygonsDataFrame(spPolygons,match.ID=T,data=data)
}
sp_dat = county_df$id %>%
unique() %>%
factor() %>%
data.frame(name=.)
rownames(sp_dat) = sp_dat$name
sp_dat$id = as.numeric(sp_dat$name)
counties <- points2polygons(county_df, sp_dat) #this may take a moment
locs = counties$name %>%
as.character() %>%
strsplit(split = ",_") %>%
do.call("rbind", .)
counties$county = locs[,1]
counties$state = locs[,2]Once completed, I had a map of the United States composed of polygons for each county. Now, any new columns added to d (an ‘sf’ object) will be automatically georeferenced.
完成后,我將獲得一個由每個縣的多邊形組成的美國地圖。 現在,添加到d (“ sf”對象)的所有新列都將自動進行地理參考。
d <- st_read("raw_data/shape/counties.shp", quiet = TRUE)
st_crs(d) <- CRS("+proj=longlat")
print(d$geometry)## Geometry set for 3076 features
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: -124.6813 ymin: 25.12993 xmax: -67.00742 ymax: 49.38323
## CRS: +proj=longlat +ellps=WGS84
## First 5 geometries:
## MULTIPOLYGON (((-86.50517 32.3492, -86.53382 32...
## MULTIPOLYGON (((-87.93757 31.14599, -87.93183 3...
## MULTIPOLYGON (((-85.42801 31.61581, -85.74313 3...
## MULTIPOLYGON (((-87.02083 32.83621, -87.30731 3...
## MULTIPOLYGON (((-86.9578 33.8618, -86.94062 33....d %>%
ggplot() +
theme_minimal() +
geom_sf(fill="steelblue", color="white", size=0.1)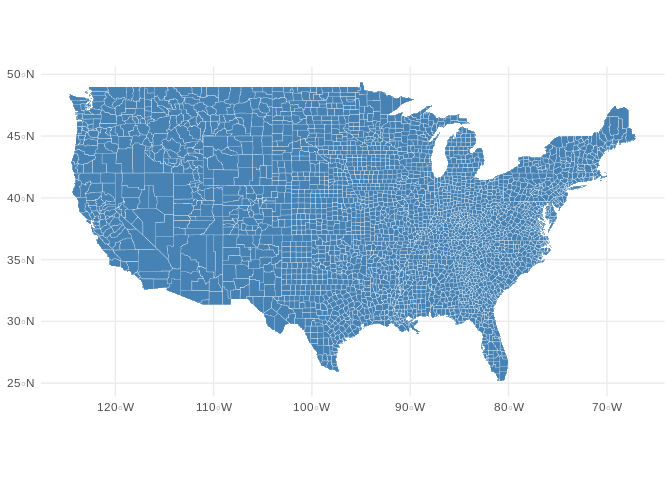
1. WorldClim的氣候變量 (1. Climatic Variables from WorldClim)
To characterize the climatic conditions for each county, I turned to WorldClim’s global climate database, specifically their ‘bioclim’ collection of annual climate summary measurements. As with the county-level polygons, there exists an API for downloading these files directly from R, this time using the raster::getData() function. Below is the code for downloading ‘bioclim’ data at a resolution of 5 arc minutes, or approximately 64 km2 per grid cell at the latitudes considered here.
為了描述每個縣的氣候條件,我求助于WorldClim的全球氣候數據庫,特別是他們的“ bioclim”年度氣候摘要測量值集合。 與縣級多邊形一樣,有一個API可直接從R下載這些文件,這次使用raster::getData()函數。 以下是在此處考慮的緯度下,以5弧分鐘的分辨率或每個網格單元大約64km2的分辨率下載“ bioclim”數據的代碼。
##Download environmental variables
getData('worldclim', var='bio', res=2.5, path = "raw_data/") #123 MB download
varsToGet = c("tmean","tmin","tmax","prec","bio","alt")
sapply(varsToGet, function(x) getData(name = "worldclim", download = TRUE, res=10, var=x))#bioclim variables (https://worldclim.org/raw_data/bioclim.html)
# BIO1 = Annual Mean Temperature
# BIO2 = Mean Diurnal Range (Mean of monthly (max temp - min temp))
# BIO3 = Isothermality (BIO2/BIO7) (×100)
# BIO4 = Temperature Seasonality (standard deviation × 100)
# BIO5 = Max Temperature of Warmest Month
# BIO6 = Min Temperature of Coldest Month
# BIO7 = Temperature Annual Range (BIO5-BIO6)
# BIO8 = Mean Temperature of Wettest Quarter
# BIO9 = Mean Temperature of Driest Quarter
# BIO10 = Mean Temperature of Warmest Quarter
# BIO11 = Mean Temperature of Coldest Quarter
# BIO12 = Annual Precipitation
# BIO13 = Precipitation of Wettest Month
# BIO14 = Precipitation of Driest Month
# BIO15 = Precipitation Seasonality (Coefficient of Variation)
# BIO16 = Precipitation of Wettest Quarter
# BIO17 = Precipitation of Driest Quarter
# BIO18 = Precipitation of Warmest Quarter
# BIO19 = Precipitation of Coldest QuarterWith the raw climate data saved to a local directy, the *.bil files are identified, sorted by name, and queued for import back into the R environment.
將原始氣候數據保存到本地目錄后,*。bil文件被標識,按名稱排序,并排隊等待導入回到R環境中。
bclimFiles = list.files("raw_data/wc5/", pattern = "bio", full.names = TRUE)[-1]
bclimFiles = bclimFiles[str_detect(bclimFiles, "bil")==TRUE]
orderFiles = str_extract_all(bclimFiles, pattern="[[:digit:]]+") %>%
do.call("rbind",.) %>%
as.matrix %>%
apply(., 2, as.numeric)
orderFiles = order(orderFiles[,2])
bioclim = stack(bclimFiles[orderFiles]) %>%
crop(., extent(d) + c(-1,1) + 2)plot(bioclim, col=viridis::viridis(15))
Feature extraction using principal components analysis
使用主成分分析進行特征提取
The dataset includes 19 variables but deal mostly with characterizing temperature and precipitation patterns. As a result, we might expect that many of these variables will be highly correlated with one another, and would want to consider using feature extraction to distill the variation to fewer dimensions.
該數據集包括19個變量,但主要處理特征溫度和降水模式。 結果,我們可能期望其中許多變量彼此高度相關,并希望考慮使用特征提取將變化提煉為更少的維度。
Indeed what I have chosen to do here is perform principal components analysis (PCA) on the climate data in order to collapse raw variables into a smaller number of axes. Before I did that, however, it was important to first normalize each variable by centering each around its mean and re-scaling the variance to 1 by dividing measurements by their standard deviation. After processing the data and performing the PCA, I found that approximately 85% of the total variation could was captured by the first four principal components, and chose to hold onto these axes of variation in order to characterize climatic variation in the data set.
確實,我在這里選擇做的是對氣候數據執行主成分分析(PCA),以將原始變量分解為較少數量的軸。 但是,在執行此操作之前,重要的是首先通過將每個變量均值居中并將每個變量除以標準差將其重新標定為1來對每個變量進行歸一化。 在處理了數據并執行了PCA之后,我發現前四個主要成分可以捕獲大約85%的總變化,并選擇保留這些變化軸以表征數據集中的氣候變化。
#normalize data to mean 0 and unit variance
normalize <- function(x){
mu <- mean(x, na.rm=TRUE)
sigma <- sd(x, na.rm=TRUE)
y <- (x-mu)/sigma
return(y)
}i = 1:19
vals <- getValues(bioclim[[i]])
completeObs <- complete.cases(vals)
vals2 <- apply(vals, 2, normalize)pc <- princomp(vals2[completeObs,], scores = TRUE, cor = TRUE)
pc$loadings[,1:4]## Comp.1 Comp.2 Comp.3 Comp.4
## bio1 0.34698924 0.037165967 0.18901924 0.05019798
## bio2 0.15257694 -0.272739207 -0.07407268 0.03206231
## bio3 0.31815555 -0.074479020 -0.21362231 -0.04928870
## bio4 -0.31781820 -0.057012175 0.27166055 -0.02437292
## bio5 0.28898841 -0.097911139 0.29797222 0.10820004
## bio6 0.35539295 0.070110518 -0.01124831 0.07097175
## bio7 -0.28661480 -0.164457842 0.22300486 -0.02206691
## bio8 0.13432442 -0.025862291 0.53410866 -0.22309867
## bio9 0.30712207 0.051220831 -0.19414056 0.22068531
## bio10 0.28407612 0.006029582 0.38463344 0.07113897
## bio11 0.36260850 0.039790805 0.02402410 0.05285433
## bio12 -0.01075743 0.398284654 0.01977560 -0.06418026
## bio13 0.06108500 0.334409706 -0.05634189 -0.40179441
## bio14 -0.05732622 0.345276698 0.11795006 0.32544197
## bio15 0.15609901 -0.200167882 -0.05441272 -0.56882750
## bio16 0.03787519 0.343597936 -0.07294893 -0.38402529
## bio17 -0.04715319 0.356553009 0.10933013 0.30589465
## bio18 -0.03660802 0.280740110 0.34421087 -0.18227552
## bio19 0.03273085 0.339449046 -0.27023885 -0.02370295pcPredict <- predict(pc, vals)r1 = r2 = r3 = r4 = raster(bioclim)
r1[] = pcPredict[,1]
r2[] = pcPredict[,2]
r3[] = pcPredict[,3]
r4[] = pcPredict[,4]
pcClim = stack(r1,r2,r3,r4)
saveRDS(pcClim, file = "raw_data/bioclim_pca_rasters.rds")pcClim = readRDS("raw_data/bioclim_pca_rasters.rds")
names(pcClim) = c("CLIM_PC1","CLIM_PC2","CLIM_PC3","CLIM_PC4")
par(mfrow=c(2,2))
plot(pcClim, col=viridis::viridis(15))
Finally, we can extract the principal component values from the county-level polygons.
最后,我們可以從縣級多邊形中提取主成分值。
bioclim_ex = raster::extract(
pcClim,
d,
fun = mean
)d = cbind(d, bioclim_ex)2.人口,政治和經濟指標 (2. Demographic, Political, and Economic Indicators)
The data for demographic, political, and economic variables come from a combination of two different county level data sources. The MIT Election Lab provides the ‘election-context-2018’ data set via their GitHub page, which includes information on the outcomes of state and national elections in addition to an impressive variety of demographic variables for each county (e.g, total population, median household income, education, etc.). Though extensive, I chose to complement these data with poverty rate estimates provided by the U.S. Census Bureau. Combining these data tables with the ‘master’ data table object (d) was a cinch using the dplyr::left_join() function.
人口,政治和經濟變量的數據來自兩個不同縣級數據源的組合。 麻省理工學院選舉實驗室通過其GitHub頁面提供了'election-context-2018'數據集,該數據集包含有關州和全國選舉結果的信息,以及每個縣令人印象深刻的各種人口統計學變量(例如,總人口,中位數)家庭收入,教育程度等)。 盡管內容廣泛,但我還是選擇了美國人口普查局提供的貧困率估算值來補充這些數據。 使用dplyr::left_join()函數可以將這些數據表與“主”數據表對象( d )結合起來。
countyDemo = "raw_data/2018-elections-context.csv" %>%
read.csv(stringsAsFactors = FALSE) %>%
mutate(state = tolower(state)) %>%
mutate(county = tolower(county)) %>%
mutate(county = str_replace(county, "st ", "saint ")) %>%
mutate(county = str_replace(county, "ste ", "saint ")) %>%
mutate(state = str_replace(state, "\\s", "_")) %>%
mutate(county = str_replace(county, "\\s", "_")) %>%
mutate(name = paste(county, state, sep=",_")) %>%
filter(!duplicated(name)) %>%
mutate(total_population = log(total_population)) %>%
mutate(white_pct = asin(white_pct/100)) %>%
mutate(black_pct = asin(black_pct/100)) %>%
mutate(hispanic_pct = asin(hispanic_pct/100)) %>%
mutate(foreignborn_pct = asin(foreignborn_pct/100)) %>%
mutate(age29andunder_pct = asin(age29andunder_pct/100)) %>%
mutate(age65andolder_pct = asin(age65andolder_pct/100)) %>%
mutate(rural_pct = asin(rural_pct/100)) %>%
mutate(lesshs_pct = asin(lesshs_pct/100))countyPoverty = "raw_data/2018-county-poverty-SAIPESNC_25AUG20_17_45_39_40.csv" %>%
read.csv(stringsAsFactors = FALSE) %>%
mutate(Median_HH_Inc = str_remove_all(Median.Household.Income.in.Dollars, "[\\$|,]")) %>%
mutate(Median_HH_Inc = as.numeric(Median_HH_Inc)) %>%
mutate(Median_HH_Inc = log(Median_HH_Inc)) %>%
mutate(poverty_pct = asin(All.Ages.in.Poverty.Percent/100))countyAll = countyDemo %>%
left_join(countyPoverty, by=c("fips"="County.ID"))#Derived variables
#2016 Presidential Election
totalVotes2016 = cbind(countyDemo$trump16,countyDemo$clinton16, countyDemo$otherpres16) %>% rowSums()
countyAll$trump16_pct = (countyDemo$trump16 / totalVotes2016) %>% asin()#2012 Presidential Election
totalVotes2012 = cbind(countyDemo$obama12,countyDemo$romney12, countyDemo$otherpres12) %>% rowSums()
countyAll$romney12_pct = (countyDemo$romney12 / totalVotes2012) %>% asin()social_vars = c("total_population","white_pct","black_pct","hispanic_pct","foreignborn_pct",
"age29andunder_pct","age65andolder_pct","rural_pct")economic_vars = c("Median_HH_Inc","clf_unemploy_pct","poverty_pct","lesscollege_pct","lesshs_pct")political_vars = c("trump16_pct","romney12_pct")all_vars = c(social_vars,economic_vars,political_vars)d <- d %>%
left_join(countyAll[, c("name", all_vars)], by = "name")## Warning: Column `name` joining factor and character vector, coercing into
## character vectorggplot(d) +
theme_minimal() +
ggtitle("Population by U.S. County") +
geom_sf(mapping=aes(fill=total_population), size=0.1, color="gray50") +
scale_fill_viridis_c("(log scale)")
3.公共土地和保護區 (3. Public Land and Protected Areas)
Both my parents and myself enjoy the outdoors, and are accustomed to having plenty of public lands nearby for a day hike, camping trip, or mountain bike ride.
我的父母和我自己都喜歡戶外活動,并且習慣于在附近有很多公共土地來進行一日遠足,露營旅行或騎山地自行車。
The International Union for the Conservation of Nature (IUCN) makes available to the public a wonderful geospatial data set covering over 33,000 protected areas in the United States alone. Covering everything from small local nature preserves and city parks to iconic National Parks, the data set is as extensive as it is impressive, but this level of detail also makes it computationally burdensome to work with. Therefore, my first steps in processing this data were removing regions of the world not considered here as well as those protected land categories unrelated to recreation.
國際自然保護聯盟(IUCN)向公眾提供了精彩的地理空間數據集,僅在美國就覆蓋了33,000多個保護區。 數據集涵蓋了從小型本地自然保護區和城市公園到標志性的國家公園在內的所有內容,其數據范圍之廣令人印象深刻,但是細節水平也使計算工作負擔沉重。 因此,我處理這些數據的第一步是刪除此處未考慮的世界區域以及與娛樂無關的受保護土地類別。
# https://www.iucn.org/theme/protected-areas/about/protected-area-categories
##Subset / clean data
#read into workspace
wdpa = readOGR("raw_data/WDPA/WDPA_Apr2020_USA-shapefile-polygons.shp")
#PAs in contiguous United States
rmPA = c("US-AK","US-N/A;US-AK","US-N/A","US-HI","US-N/A;US-AK","US-N/A;US-CA","US-N/A;US-FL","US-N/A;US-FL;US-GA;US-SC","US-N/A;US-HI")
wdpa_48 = wdpa[!wdpa$SUB_LOC %in% rmPA,]
#Only terrestrial parks
wdpa_48_terr = wdpa_48[wdpa_48$MARINE==0,]
#Subset by PA type
paLevels = c("Ib","II","III","V","VI")
wdpa_48_terr_pa = wdpa_48_terr[wdpa_48_terr$IUCN_CAT %in% paLevels,]
writeSpatialShape(wdpa_48_terr_pa, fn = "raw_data/WDPA/Derived/public_lands_recreation.shp", factor2char = TRUE)After paring the data down, the intermediate product is saved to local storage so that I the process would not have to be repeated when re-running this code. From this, the proportional representation of protected land within each county is calculated. With over 3,000 counties in the United States, I knew that this process would take some computing time. In order to speed things up, I decided to parallelize the task with the foreach and doMC packages. Please note that if you are implementing this code on your own machine, you may need to find substitute packages that are compatible with your operating system. Operating systems require different back ends to allow parallelization. The code here was implemented on a Linux machine.
縮減數據后,中間產品將保存到本地存儲中,這樣在重新運行此代碼時不必重復該過程。 據此計算出每個縣內保護地的比例表示。 我知道在美國有3,000多個縣,這一過程將需要一些計算時間。 為了加快速度,我決定將任務與foreach和doMC軟件包并行化。 請注意,如果要在自己的計算機上實現此代碼,則可能需要查找與您的操作系統兼容的替代軟件包。 操作系統需要不同的后端以允許并行化。 這里的代碼是在Linux機器上實現的。
pa_by_county <- readRDS("pa_by_county.rds")#add data to sf object
vars = names(pa_by_county)[-which(colSums(pa_by_county)==0)]
for(v in 1:length(vars)){
d[vars[v]] <- pa_by_county[[vars[v]]] %>% asin()
}ggplot(d) +
theme_minimal() +
ggtitle("Proportion of Protected Land by U.S. County") +
geom_sf(mapping=aes(fill=boot::logit(TOT_PA_AREA+0.001)), size=0.1) +
scale_fill_viridis_c("logit(%)")
4.國家土地覆蓋數據集(2016年) (4. National Land Cover Dataset (2016))
The National Land Cover Database, a high resolution (30 m x 30 m grid cells) land cover category map produced by the USGS and other federal agencies. Each cell on the map corresponds to one of 16* land cover categories including bare ground, medium intensity development, deciduous forests, cultivated land, and more. The NLCD data is very much integral to the design of Niche because it provides a powerful way of quantifying some of the most salient characteristics of a location: what lies on top of the land. Maybe your ideal place to live has evergreen forests, small towns, and large expanses of grazing pasture. Perhaps it consists mainly of high intensity development, such as in a metro area, and with little or absent forest and agriculture land uses. The NLCD2016 data can capture represent these characteristics using combinations of these categories.
國家土地覆蓋數據庫 ,由USGS和其他聯邦機構制作的高分辨率(30 mx 30 m網格單元)土地覆蓋類別圖。 地圖上的每個單元格對應16種土地覆蓋類別之一,包括裸露土地,中等強度的開發,落葉林,耕地等 。 NLCD數據對于Niche的設計非常重要,因為它提供了一種強大的方法來量化某個位置的一些最突出的特征:位于地面上的東西。 也許您理想的居住地是常綠森林,小鎮和廣闊的牧場。 也許它主要由高強度開發組成,例如在大都市地區,幾乎沒有森林和農業用地。 NLCD2016數據可以使用這些類別的組合來捕獲以代表這些特征。
As with other spatial data extractions, the goal was to iterate over counties in the data set and calculate the proportional representation of each land cover category in that county, and it was here that I ran into some stumbling blocks. The root of these issues was a combination of the sheer size of some U.S. counties and the high spatial resolution of the NLCD2016 data. Initially, I tried to use the raster::extract() pull out a vector of values from the cells falling within them, and, as before, to parallelize this task across CPU cores. This worked fine at first, but I quickly encountered large spikes in system memory use followed by crashes of my operating system. The problem was that some large counties (like San Bernardino County, California) are enormous and encompass areas far larger than many U.S. states. Attempting to hold all this data in system memory was just not going to work.
與其他空間數據提取一樣,目標是迭代數據集中的各個縣,并計算該縣每個土地覆蓋類別的比例表示形式,正是在這里,我遇到了一些絆腳石。 這些問題的根源是一些美國縣的龐大規模和NLCD2016數據的高空間分辨率的結合。 最初,我嘗試使用raster::extract()從屬于它們的單元格中raster::extract()值的向量,并且像以前一樣,跨CPU內核并行執行此任務。 起初效果不錯,但是我很快就遇到了系統內存使用量的大峰值,然后操作系統崩潰了。 問題在于,一些大縣(例如加利福尼亞的圣貝納迪諾縣)規模巨大,而且所涵蓋的地區遠遠超過美國許多州。 試圖將所有這些數據保存在系統內存中只是行不通。
Next, I tried cropping the NLCD2016 map to the spatial extent of the county before attempting to extract data. Operations were indeed much speedier than before, but before long I encountered the same system memory issues as before.
接下來,在嘗試提取數據之前,我嘗試將NLCD2016地圖裁剪到該縣的空間范圍。 操作確實比以前快得多,但是不久之后,我遇到了與以前相同的系統內存問題。
My eventual workaround involved dividing the spatial polygons for large counties into smaller subsets, and then sequentially extracting the map cell values using these sub-polygons in order to avoid a surge in memory use. The results extracted from each polygon were then written to storage in JSON format, under a directory containing all other files for that county. Later, this data can be retrieved and combined with the other files to reconstruct the land cover classes in the entire county. Even still, the entire process took quite some time to complete. On my computer, it took over 16 hours to complete once I had the code worked out.
我最終的解決方法是將大型縣的空間多邊形分成較小的子集,然后使用這些子多邊形順序提取地圖單元的值,以避免內存使用激增。 然后將從每個多邊形中提取的結果以JSON格式寫入到存儲中,該目錄包含該縣的所有其他文件。 之后,可以檢索此數據并將其與其他文件組合以重建整個縣的土地覆蓋類別。 即便如此,整個過程還是需要花費一些時間才能完成。 在計算機上完成代碼處理后,花了16個小時以上才能完成。

Here is the full code, with some added conditional statements to check whether files with the same data already exist in storage.
這是完整的代碼,并添加了一些條件語句以檢查存儲中是否已存在具有相同數據的文件。
library(foreach)
library(doMC)s = Sys.time()
registerDoMC(cores = 6)
index = 1:length(counties2)
county_state <- sapply(counties2, function(x)
x$name) %>%
as.character()foreach(i = index) %dopar% {
#for(i in index){
name. <- county_state[i]
fileName <-
paste0(formatC(i, width = 4, flag = "0"), "_", name.)
dirName <- sprintf("land_use/%s", fileName)
if (!dir.exists(dirName)) {
dir.create(dirName)
}
fileOut <- sprintf("%s/%s.json", dirName, fileName)
if (!file.exists(fileOut)) {
#countyPoly = counties2[[i]]
countyPoly = gBuffer(counties2[[i]], byid = TRUE, width = 0) #fixes topology error (RGEOS)
area. <- (gArea(countyPoly) * 1e-6)
if (area. > 1e3) {
dims <-
ifelse(area. > 2.5e4,
6,
ifelse(
area. <= 2.5e4 & area. > 1e4,
4,
ifelse(area. > 1e3 & area. <= 1e4, 2, 1)
))
grd <-
d[d$name == name., ] %>%
st_make_valid() %>%
sf::st_make_grid(n = dims) %>%
st_transform(., crs = st_crs(p4s_nlcd)) %>%
as(., 'Spatial') %>%
split(., .@plotOrder)
grd_list <- lapply(grd, function(g)
crop(countyPoly, g))
sapply(1:length(grd_list), function(j) {
fileName2 <- str_split(fileName, "_", n = 2) %>%
unlist()
fileName2 <-
paste0(fileName2[1], paste0(".", j, "_"), fileName2[2])
fileOut2 <- sprintf("%s/%s.json", dirName, fileName2)
if (!file.exists(fileOut2)) {
countyPoly = gBuffer(grd_list[[j]], byid = TRUE, width = 0) #fixes topology error (RGEOS)
nlcd_sub <- crop(nlcd, grd.)
cellTypes <- raster::extract(nlcd_sub, grd.) %>%
unlist()
yf <- factor(cellTypes, landUseLevels) %>%
table() %>%
#prop.table() %>%
data.frame()
names(yf) <- c("category", "Count")
jsonlite::write_json(yf, path = fileOut2)
}
})
}
else{
nlcd2 <- crop(nlcd, countyPoly)
cellTypes <- raster::extract(nlcd2, countyPoly) %>%
unlist()
yf <- factor(cellTypes, landUseLevels) %>%
table() %>%
#prop.table() %>%
data.frame()
names(yf) <- c("category", "Count")
jsonlite::write_json(yf, path = fileOut)
}
}
}The data from JSON files can be easily retrieved by iterating a custom ‘ingestion’ function over the directories for each county. As with the climate data, we want to consider if feature extraction with PCA might be appropriate here given the large number of land cover classes featured in the data set, the presence or absence of which are likely to be correlated with one another.
通過在每個縣的目錄上迭代自定義“輸入”功能,可以輕松檢索JSON文件中的數據。 與氣候數據一樣,我們要考慮的是,鑒于數據集中有大量的土地覆蓋類別,而有無可能相互關聯,因此在這里用PCA進行特征提取是否合適。
dirs <- list.dirs("land_use/", recursive = TRUE)[-1]#function for reading and processing the files in each directory (county)
ingest <- function(x){
f <- list.files(x, full.names = TRUE)
d <- lapply(f, function(y){
z <- read_json(y)
lapply(z, data.frame) %>%
do.call("rbind",.)
}) %>%
do.call("rbind",.)
out <- tapply(d$Count, list(d$category), sum) %>%
prop.table()
return(out)
}#iterate read and processing function over directories
lc <- lapply(dirs, ingest) %>%
do.call("rbind",.)
#remove columns for land use categories unique to Alaska
rmCols <- which(attributes(lc)$dimnames[[2]] %in% paste(72:74))#feature reduction using PCA
pc <- princomp(lc[,-rmCols], scores = TRUE, cor = FALSE)
scores <-pc$scores[,1:4] %>% data.frame()
names(scores) <- vars <- paste0("LandCover_PC",1:4)
par(mfrow=c(2,2))
for(v in 1:length(vars)){
d[vars[v]] <- scores[[vars[v]]]
}library(ggplotify)
library(gridExtra)p1 <- ggplot(d) +
theme_minimal() +
ggtitle("Land Cover PC1") +
geom_sf(mapping=aes(fill=LandCover_PC1), size=0.1) +
scale_fill_viridis_c("")p2 <- ggplot(d) +
theme_minimal() +
ggtitle("Land Cover PC2") +
geom_sf(mapping=aes(fill=LandCover_PC2), size=0.1) +
scale_fill_viridis_c("")plt <- gridExtra::arrangeGrob(grobs = lapply(list(p1,p2), as.grob), ncol=1)
grid::grid.newpage()
grid::grid.draw(plt)
5.提出建議:余弦接近 (5. Making recommendations: cosine proximity)
With all of the data in place, it is now time to put the pieces together and make some recommendations about where to potentially move. This works by calculating the distance between the focal county (the basis of the comparison) and all others in the country using the cosine similarity measure. Before getting more into that, the features to include in that calculation need to be isolated and some transformations performed.
有了所有數據之后,現在就可以將各個部分放在一起,并對可能的移動位置提出一些建議。 通過使用余弦相似性度量來計算重點縣(比較的基礎)與該國所有其他縣之間的距離,可以實現此目的。 在深入探討之前,需要將要包括在該計算中的功能隔離開并進行一些轉換。
climate_vars = paste0("CLIM_PC", 1:3)social_vars = c(
"total_population",
"white_pct",
"black_pct",
"hispanic_pct",
"foreignborn_pct",
"age29andunder_pct",
"age65andolder_pct",
"rural_pct"
)economic_vars = c(
"Median_HH_Inc",
"clf_unemploy_pct",
"poverty_pct",
"lesscollege_pct",
"lesshs_pct"
)political_vars = c("trump16_pct",
"romney12_pct"
)landscape_vars = paste0("LandCover_PC", 1:4)features = c(climate_vars,
social_vars,
economic_vars,
political_vars,
landscape_vars)
## Variable transformation function
transform_data <- function(x) {
min_x = min(x, na.rm = TRUE)
max_x = max(x, na.rm = TRUE)
mean_x = mean(x, na.rm = TRUE)
y = (x - mean_x) / (max_x - min_x)
return(y)
}
vals <- d[,features]
vals$geometry <- NULL
vals <- apply(vals, 2, transform_data)
vals_t <- t(vals)
attr(vals_t, "dimnames") <- NULLCosine similarity is a common method used in recommender systems to identify similar entities (books, movies, etc.) on the basis of a vector of data features. In the previous block of code, we created and (N x M) matrix where N is the number of features in the data set and M is the number of counties. Cosine similarity works by calculating the angle (θ?,?) from the edges formed between the column vector of the focal county (m?) and a comparison county (m?) in n-dimensional space. The cosine of two vectors that share many properties will have an angle close to 0 and therefore a cosine value near 1. Conversely, dissimilar counties will have lower scores because the angle of their edge will be larger. The formula for calculating cosine similarity is as follows:
余弦相似度是推薦系統中基于數據特征向量識別相似實體(書籍,電影等)的常用方法。 在上一個代碼塊中,我們創建了(N x M)矩陣,其中N是數據集中的要素數量,M是縣數量。 余弦相似度是通過計算n維空間中焦點縣(m?)和比較縣(m?)的列向量之間形成的邊緣的角度(θ?,?)來進行的。 具有許多特性的兩個向量的余弦將具有接近0的角度,因此余弦值接近1。相反,相異的縣的得分較低,因為它們的邊緣角度會更大。 余弦相似度的計算公式如下:
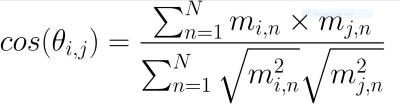
The code for this calculation is given below along with an example for Cook County, Illinois, which includes the city of Chicago.
下面給出了此計算的代碼,并提供了一個伊利諾伊州庫克縣的示例,其中包括芝加哥市。
cosine_similarity <- function(x, focal_index, measure = "angle") {
assertthat::assert_that(measure %in% c("angle", "distance"),
length(measure) == 1)
cos_prox = rep(NA, ncol(x))
for (i in 1:ncol(x)) {
if (focal_index != i) {
A = x[, focal_index] %>%
as.matrix(ncol = 1)
A_norm = (A * A) %>% sum() %>% sqrt()
B = x[, i] %>%
as.matrix(ncol = 1)
B_norm = (B * B) %>% sum() %>% sqrt()
dot_AB = sum(A * B)
cos_prox[i] = dot_AB / (A_norm * B_norm)
}
}
if (measure=="angle")
return(cos_prox)
else
cos_distance = acos(cos_prox) / pi
return(cos_distance)
}
#Measure cosine distance among counties, relative to focal county
focal_location <- "cook,_illinois"
focal_index <-
which(d$name == focal_location) #index of focal location
d$cosine_sim <- cosine_similarity(vals_t, focal_index, measure = "angle")#Top 10 Recommendations
d[order(d$cosine_sim,decreasing = TRUE),c("name","cosine_sim")]## Simple feature collection with 3076 features and 2 fields
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: -124.6813 ymin: 25.12993 xmax: -67.00742 ymax: 49.38323
## CRS: +proj=longlat +ellps=WGS84
## First 10 features:
## name cosine_sim geometry
## 1194 suffolk,_massachusetts 0.9367209 MULTIPOLYGON (((-71.17855 4...
## 710 marion,_indiana 0.9346767 MULTIPOLYGON (((-86.31609 3...
## 1816 kings,_new_york 0.9300806 MULTIPOLYGON (((-73.86572 4...
## 3018 milwaukee,_wisconsin 0.9080851 MULTIPOLYGON (((-88.06934 4...
## 1740 atlantic,_new_jersey 0.9072181 MULTIPOLYGON (((-74.98299 3...
## 1852 westchester,_new_york 0.8982588 MULTIPOLYGON (((-73.80843 4...
## 279 hartford,_connecticut 0.8946008 MULTIPOLYGON (((-72.74845 4...
## 2259 philadelphia,_pennsylvania 0.8916324 MULTIPOLYGON (((-74.97153 4...
## 217 arapahoe,_colorado 0.8858975 MULTIPOLYGON (((-104.6565 3...
## 2253 monroe,_pennsylvania 0.8852190 MULTIPOLYGON (((-74.98299 4...The Top 10 recommendations for Cook County residents are presented here. Most similar to Cook County are Suffolk County, MA, a suburb of New York City, and Marion County, IN, which contains the city of Indianapolis.
這里介紹了對庫克縣居民的十大建議。 與庫克縣最相似的是位于紐約市郊區的馬薩諸塞州薩福克縣和包含印第安納波利斯市的印第安納州馬里恩縣。
結論 (Conclusion)
This was a quick overview of the data and methods that went into Niche: An Interface for Exploring Relocation Options. If you would like to give a try, visit the most up-to-date version on my Shinyapps page (https://ericvc.shinyapps.io/Niche/) or clone the GitHub directory and run the app from your computer with RStudio. Note, the app.R file contains code for initializing a Python virtual environment (using the reticulate package), which Niche requires in order to function completely. This code causes issues when executed on my local system. Commenting out the code fixes the issue, but are otherwise required when running the hosted version of the program. Some features will not be available without the ability to execute Python code.
這是Niche:探索重定位選項的接口所使用的數據和方法的快速概述。 如果您想嘗試一下,請訪問我的Shinyapps頁面上的最新版本(https://ericvc.shinyapps.io/Niche/)或克隆GitHub目錄,然后使用RStudio從計算機上運行該應用程序。 請注意, app.R文件包含用于初始化Python虛擬環境(使用reticulate軟件包)的代碼,Niche需要此代碼才能完全運行。 在我的本地系統上執行時,此代碼會導致問題。 注釋掉該代碼可解決此問題,但在運行程序的托管版本時則需要這樣做。 如果沒有執行Python代碼的功能,某些功能將不可用。
The purpose of Niche is to help guide users’ relocation research by translating their personal preferences and past experiences into a data-centric model in order to make recommendations. Some future plans for this tool include adding historical data that would allow users to find locations in the present that are similar to a location and time from the recent past (e.g., within the last 20 years or so).
Niche的目的是通過將用戶的個人偏好和過去的經驗轉化為以數據為中心的模型來提出建議,從而幫助指導用戶進行搬遷研究。 該工具的一些未來計劃包括添加歷史數據,該歷史數據將允許用戶查找與最近的過去(例如,最近20年左右)的地點和時間相似的地點。
翻譯自: https://medium.com/@ericvc2/niche-an-interface-for-exploring-your-moving-options-858a0baa29b2
ajax不利于seo
本文來自互聯網用戶投稿,該文觀點僅代表作者本人,不代表本站立場。本站僅提供信息存儲空間服務,不擁有所有權,不承擔相關法律責任。 如若轉載,請注明出處:http://www.pswp.cn/news/389712.shtml 繁體地址,請注明出處:http://hk.pswp.cn/news/389712.shtml 英文地址,請注明出處:http://en.pswp.cn/news/389712.shtml
如若內容造成侵權/違法違規/事實不符,請聯系多彩編程網進行投訴反饋email:809451989@qq.com,一經查實,立即刪除!

















![[原創]java獲取word里面的文本](http://pic.xiahunao.cn/[原創]java獲取word里面的文本)
