📋文章目錄
- 復現目標圖片
- 繪圖前期準備
- 繪制左側回歸線圖
- 繪制右側散點圖
- 組合拼圖 (關鍵步驟!)
?? 跟著「Nature」正刊學作圖,今天挑戰復現Nature文章中的一張組合圖–左邊為 回歸曲線、右邊為 散點圖。這種組合圖在展示相關性和分組效應時非常清晰有力。
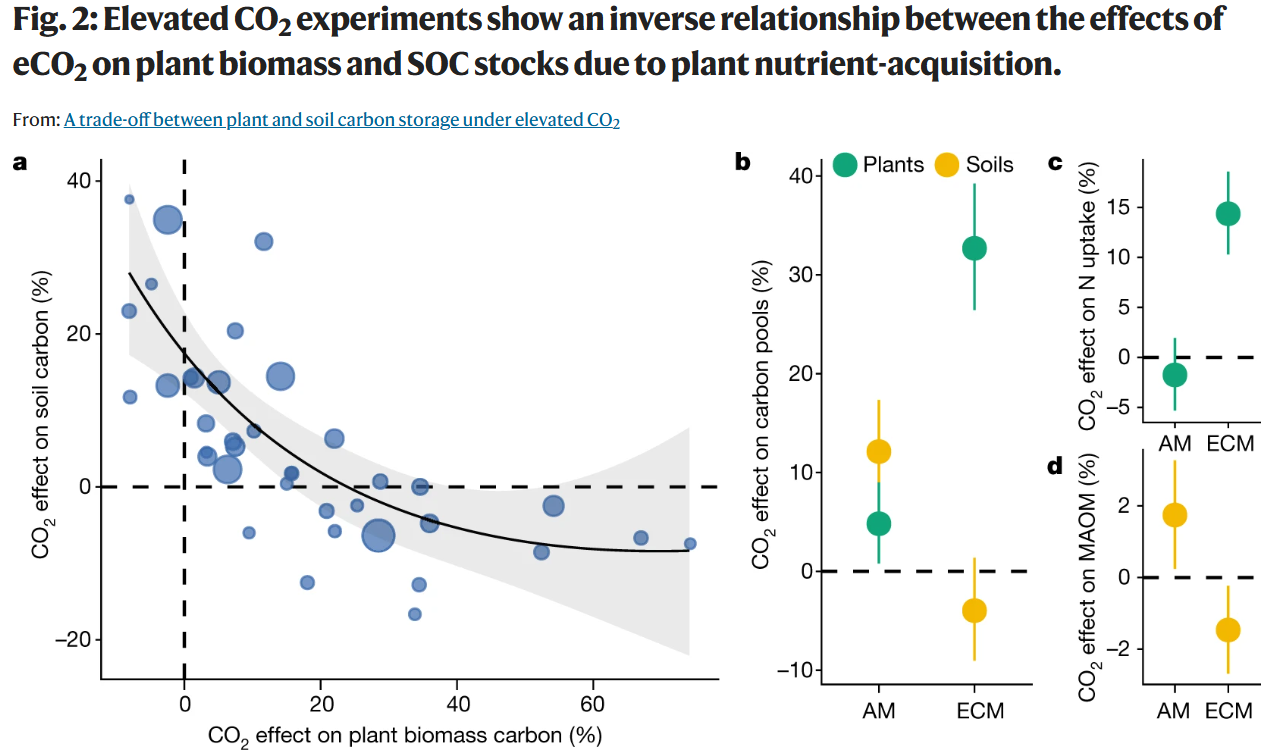
復現目標圖片

圖注:Nature原文中的組合圖 (來源 https://www.nature.com/articles/s41586-021-03306-8)
圖注:使用R ggplot2 + cowplot復現的效果
繪圖前期準備
rm(list = ls())
setwd(dirname(rstudioapi::getActiveDocumentContext()$path))library(openxlsx);library(ggplot2);library(cowplot)data<- purrr::map(1:6, ~read.xlsx("data.xlsx", .x))
# 讀取數據:6個子圖的數據分別存儲在data.xlsx的6個sheet中
data <- purrr::map(1:6, ~read.xlsx("data.xlsx", .x))
# <-- 補充說明開始 -->
# 提示:如需練習數據,可通過文末方式聯系獲取。
# 這里用map循環讀取,保證每個子圖數據獨立存儲,便于后續清晰調用。
# <-- 補充說明結束 -->
繪制左側回歸線圖
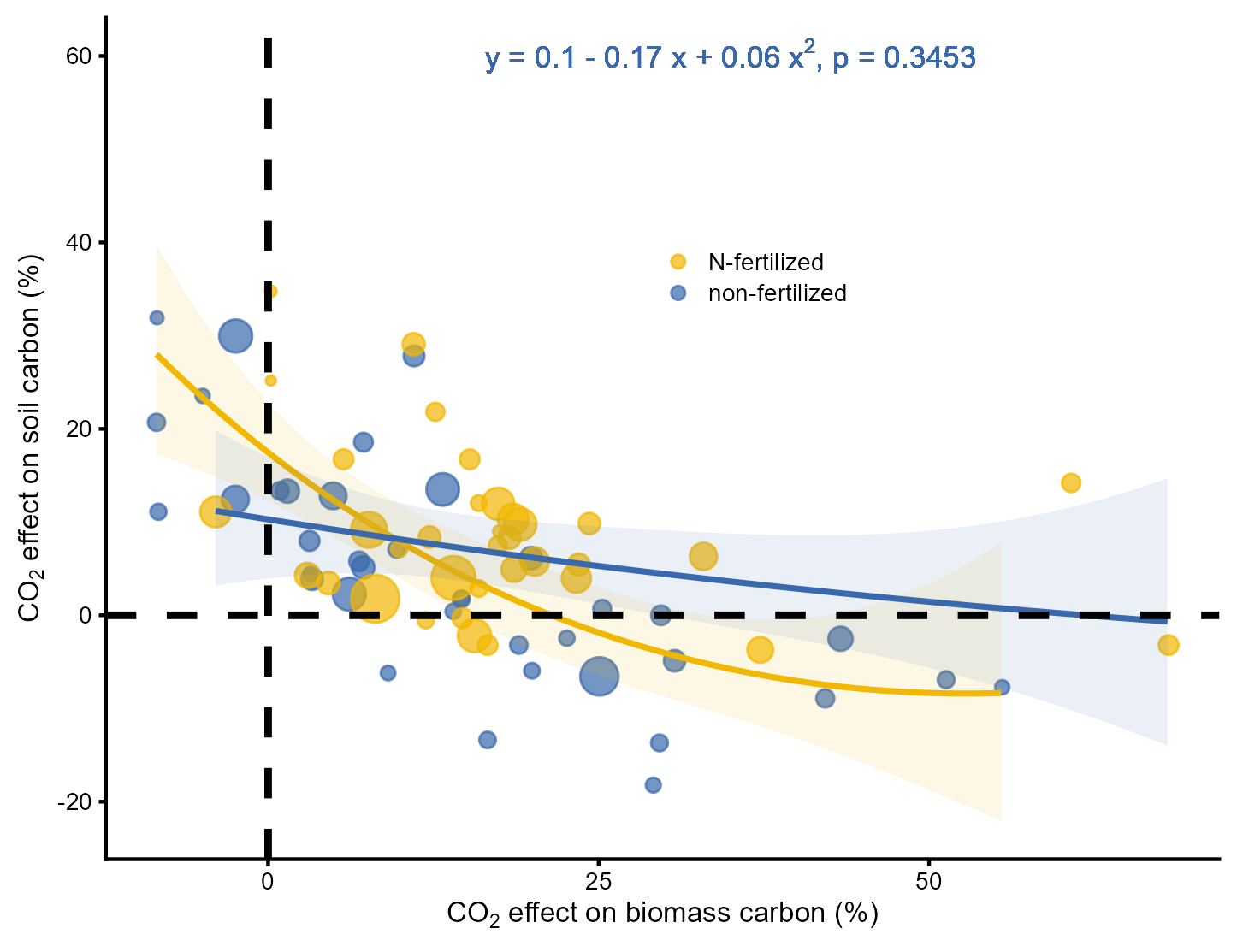
Lineplot<- ggplot(data[[1]], aes(Biomass*100, yi*100))+geom_point(aes(size = 1/vi, col = Treatment, fill = Treatment), alpha = 0.7)+#設置點的大小geom_line(aes(Biomass*100, yhat), data[[2]], col = "#F2B701", size = 0.8)+geom_ribbon(aes(x = Biomass*100, y = yhat, ymax = UCL, ymin = LCL),data[[2]], fill = "#F2B701", alpha = 0.1, size = 0.8)+#繪制置信區間geom_line(aes(Biomass*100, yhat), data[[3]], col = "#3969AC", size = 0.8)+#擬合曲線geom_ribbon(aes(x = Biomass*100, y = yhat, ymax = UCL, ymin = LCL),data[[3]], fill = "#3969AC", alpha = 0.1, size = 0.8)+scale_size(range = c(1, 6))+scale_color_manual(values = c("#F2B701","#3969AC"))+scale_fill_manual(values = c("#F2B701","#3969AC"))+geom_hline(yintercept = 0, lty=2, size = 1)+ geom_vline(xintercept = 0, lty=2, size = 1)+guides(size = "none")+theme_cowplot(font_size = 8)+#將字號設置為8theme(legend.position = c(0.5,0.7),legend.box = 'horizontal',legend.title = element_blank(),plot.margin = unit(c(5,5,5,5), "points"))+geom_text(aes(35, 60, label =(paste(expression("y = 0.1 - 0.17 x + 0.06 x"^2*", p = 0.3453")))),parse = TRUE, size = 3, color = "#3969AC")+#填入公式labs(x = expression(paste(CO[2], " effect on biomass carbon (%)")),y = expression(paste(CO[2], " effect on soil carbon (%)")))Lineplot
繪制右側散點圖
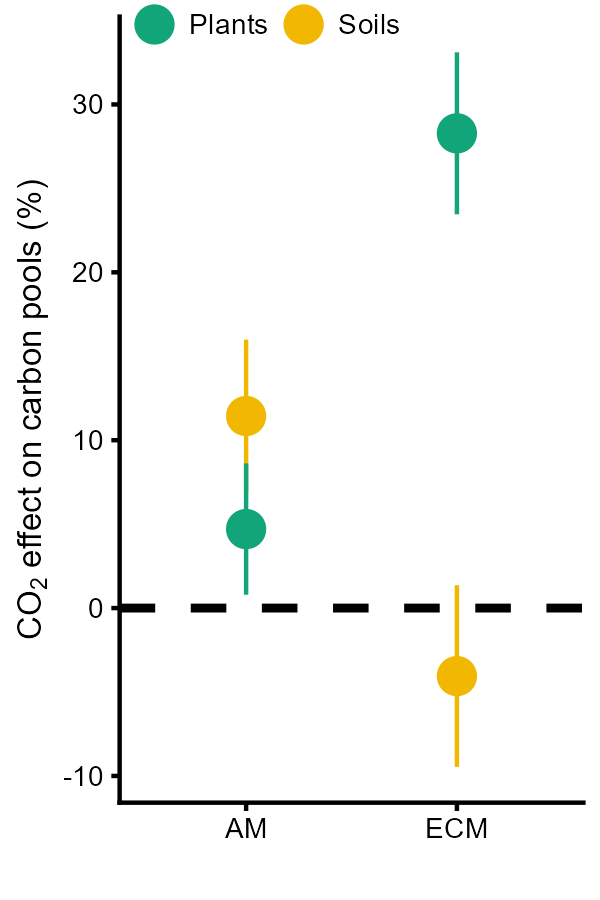
Myco<- ggplot(data[[4]], aes(Mycohiza, estimate*100, color = group, group = group))+geom_hline(yintercept = 0, lty = 2, size = 1)+ scale_color_manual(values = c("#11A579", "#F2B701"))+geom_pointrange(aes(ymin = ci.lb*100, ymax = ci.ub*100), position = position_dodge(width = 0), size = 0.8)+ theme_cowplot(font_size=8) +theme(legend.title = element_blank(),legend.direction = "horizontal",legend.position = c(0, 0.99))+labs(x = "",y = expression(paste(CO[2], " effect on carbon pools (%)")))Myco
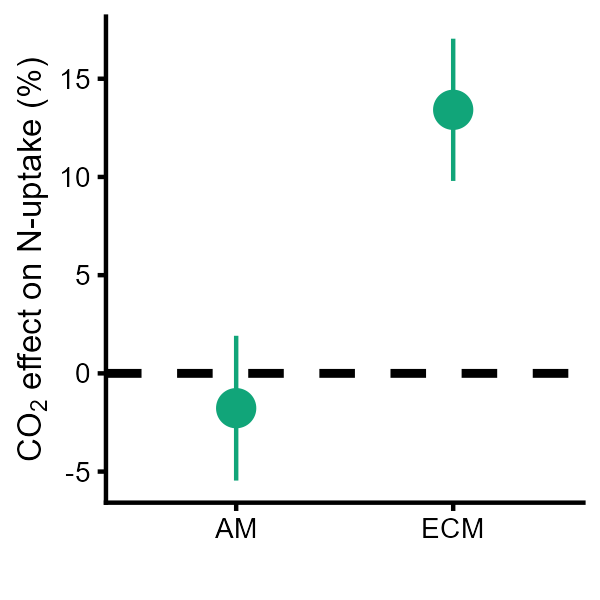
Nutake<- ggplot(data[[5]], aes(Mycohiza, estimate*100)) + geom_hline(yintercept = 0, lty=2, size=1) + geom_pointrange(aes(ymin = ci.lb*100, ymax = ci.ub*100), size = 0.8, color = "#11A579")+theme_cowplot(font_size=8) +theme(legend.position = "none",axis.title.y = element_text(margin = margin(r=1)))+labs(x = "",y = expression(paste(CO[2]," effect on N-uptake (%)")))Nutake
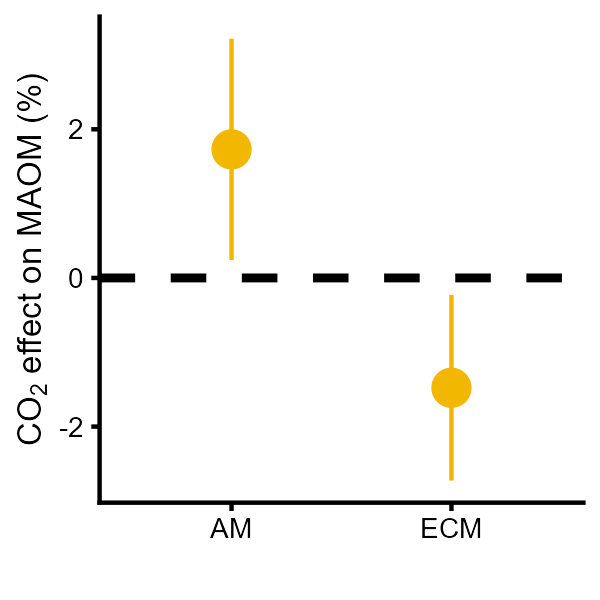
MAOM<- ggplot(data[[6]], aes(Mycohiza, estimate*100))+ geom_hline(yintercept = 0, lty = 2, size = 1)+ geom_pointrange(aes(ymin = ci.lb*100, ymax = ci.ub*100),size = 0.8, color = "#F2B701")+theme_cowplot(font_size = 8) +theme(legend.position = "none",axis.title.y = element_text(margin = margin(r=1)))+labs(x = "",y = expression(paste(CO[2]," effect on MAOM (%)")))MAOM
組合拼圖 (關鍵步驟!)
Right<- plot_grid(Nutake + theme(plot.margin = unit(c(5, 5, -10, 5), "points")),MAOM + theme(plot.margin = unit(c(0, 5, 5, 5), "points")),nrow = 2, labels = c("c","d"), align = "v", axis = "l",vjust = 1.2, hjust = 0.5, label_size = 10)#先拼接右側上下兩張圖
Midrig<- plot_grid(Myco + theme(plot.margin = unit(c(5,5,5,0), "points")),Right,vjust = 1.2,axis = "b",labels = c("b",""), label_size = 10,rel_widths = c(1, 0.7),nrow = 1, ncol = 2)#拼接所有的散點圖
Total<- plot_grid(Lineplot, middleright,vjust = 1.2, axis = "b", labels = c("a",""), label_size= 10,rel_widths = c(1, 0.7))#拼接左側的回歸曲線圖Total
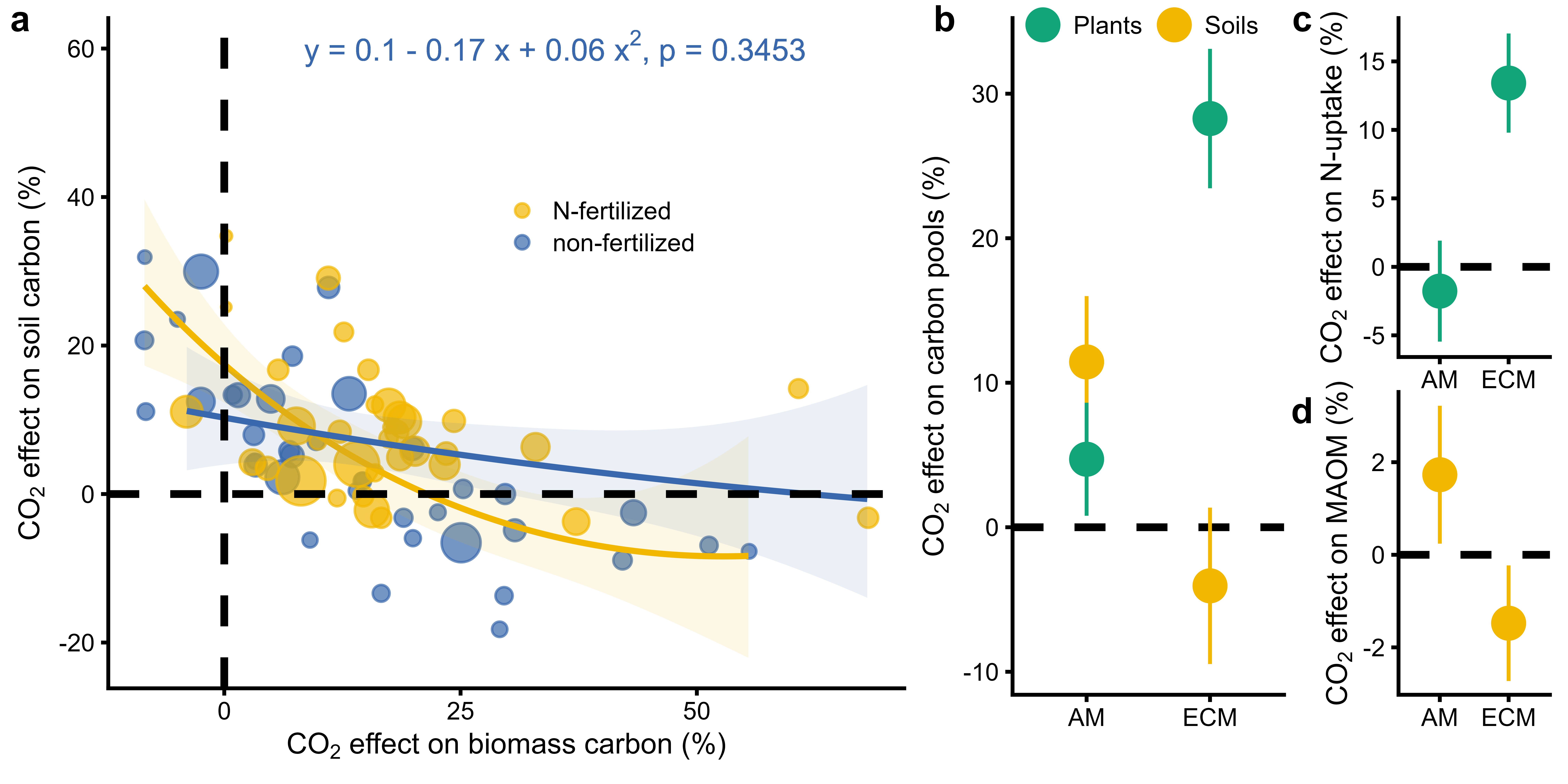
圖注:拼圖完成!關鍵點在于使用plot.margin微調子圖間距,以及rel_widths控制左右比例。
復現完成! 總結一下關鍵點:
-
數據組織:清晰分隔不同子圖所需數據。
-
回歸圖:geom_ribbon畫置信區間,size=1/vi實現加權散點。
-
點估計圖:geom_pointrange是核心,position_dodge處理分組錯位。
-
拼圖:cowplot::plot_grid是核心,精調plot.margin和rel_widths是成敗關鍵。





系統上安裝 MuJoCo)


——變分自編碼器詳解與實現)



![[AI-video] 數據模型與架構 | LLM集成](http://pic.xiahunao.cn/[AI-video] 數據模型與架構 | LLM集成)






