QIIME 2 2023.9 Amplicon Distribution介紹:
概述
qiime團隊專門針對高通量擴增子序列分析退出的conda集成環境,包括了主要和常見的擴增子分析模塊,用戶可以單獨使用各個模塊,也可以使用各模塊組成不同的分析流程。從2023.09版本開始特意將擴增子和宏基因組shotgun分成了兩個環境,可能一個環境集成太大了。這樣安裝應該更輕盈。
QIIME 2 2023.9 Amplicon Distribution
看看文章吧,超強的團隊:Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2 | Nature Biotechnology
2023.9Amplicon分發版qiime2主要功能模塊介紹:?
The 2023.9 release of the QIIME 2 Amplicon Distribution includes the QIIME 2 framework,?q2cli?(a QIIME 2 command-line interface) and the following plugins:
-
q2-alignment -
q2-composition -
q2-cutadapt -
q2-dada2 -
q2-deblur -
q2-demux -
q2-diversity -
q2-diversity-lib -
q2-emperor -
q2-feature-classifier -
q2-feature-table -
q2-fragment-insertion -
q2-longitudinal -
q2-metadata -
q2-phylogeny -
q2-quality-control -
q2-quality-filter -
q2-sample-classifier -
q2-taxa -
q2-types -
q2-vsearch
還有更多的模塊,都可以單獨安裝,一般建議成套安裝就行:
QIIME 2 Library
?
QIIME2的安裝:
還是繼續用conda吧,這個用起來也挺方便,docker稍微增加了點門檻,看個人喜好
?官方安裝介紹鏈接:
先看介紹選擇分發板,根據自己喜好安裝:Installing QIIME 2 — QIIME 2 2023.9.2 documentation

這里選擇amplicon擴增子分析的環境:
查看安裝指引:
Natively installing QIIME 2 — QIIME 2 2023.9.2 documentation
?選擇amplicon分發版的linux環境配置指導:

下載環境依賴文件并安裝:?
# 下載環境依賴配置文件
wget https://data.qiime2.org/distro/amplicon/qiime2-amplicon-2023.9-py38-linux-conda.yml# 使用mamba或conda 安裝 ,-n后的名字自己定義就行
mamba env create -n qiime2-amplicon-2023.9 --file qiime2-amplicon-2023.9-py38-linux-conda.yml要升級qiime2?,建議按日期名稱命名各個版本重新安裝,不需要的就直接刪除舊的環境,因為有些舊的環境下的模塊大家可能還需要用到,而在新的環境下可能已經廢棄或更新了,所以建議重新配置安裝新的conda環境就行。
開始使用qiime2?amplicon分發版?
要看怎么使用,或糾錯,這里應該是最全的了,User Support - QIIME 2 Forum
這里來自劉永鑫團隊的擴增子分析流程大家可以參考,把其中對應的步驟改為使用qiime模塊分析即可:
使用vsearch進行16s擴增子高通量序列分析步驟-CSDN博客
EasyAmplicon (易擴增子)-擴增子高通量序列分析軟件流程及腳本-詳細使用方法——來自劉永鑫團隊的秘籍-CSDN博客
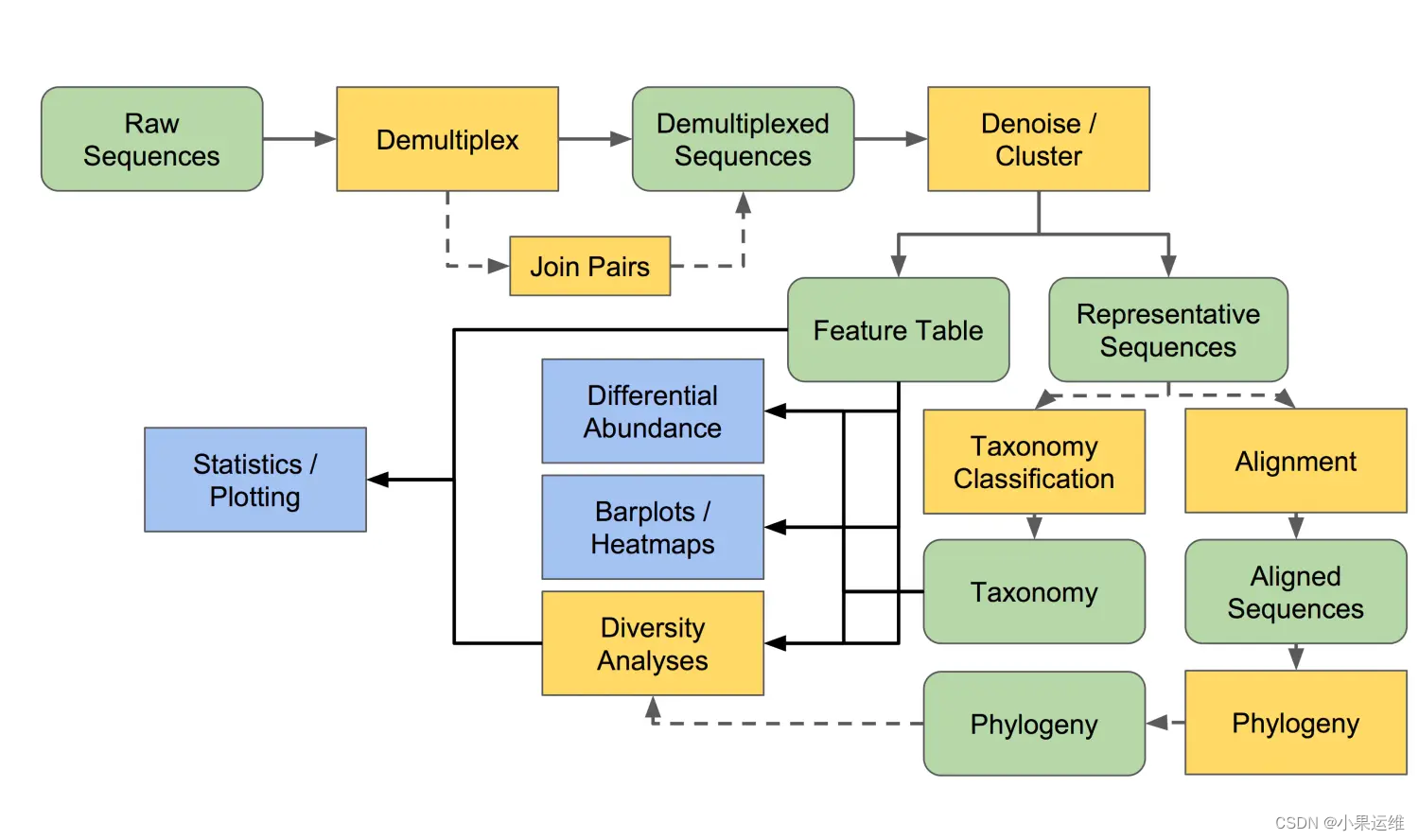
當然qiime2?不僅僅提供分析模塊,還開發了很多workflow,一個命令就可以按照workflow來得到結果了;可視化也有流程,后面大家看著參考:
?Overview of QIIME 2 Plugin Workflows — QIIME 2 2023.9.2 documentation
 ?
?
?
先激活環境并查看環境信息
# 激活環境,conda或mamba
mamba activate qiime2-amplicon-2023.9# 查看幫助信息
qiime --help

開始qiime2分析
分析前建議大家先建立獨立的工作目錄,并進入工作目錄再開始操作:
1、導入數據:
查看幫助:
![]()
qiime tools import --helpUsage: qiime tools import [OPTIONS]Import data to create a new QIIME 2 Artifact. See https://docs.qiime2.org/for usage examples and details on the file types and associated semantictypes that can be imported.Options:--type TEXT The semantic type of the artifact that will becreated upon importing. Use --show-importable-typesto see what importable semantic types are availablein the current deployment. [required]--input-path PATH Path to file or directory that should be imported.[required]--output-path ARTIFACT Path where output artifact should be written.[required]--input-format TEXT The format of the data to be imported. If notprovided, data must be in the format expected by thesemantic type provided via --type.--show-importable-types Show the semantic types that can be supplied to--type to import data into an artifact.--show-importable-formatsShow formats that can be supplied to --input-formatto import data into an artifact.--help Show this message and exit.
?大家可以先使用樣例文件做為參考,然后按樣例文件整理自己的數據
單端序列導入(序列文件與barcode文件各自獨立)
復制官方代碼時注意刪除上面的引號,這個復制過來容易變成中文的雙引號,另外還需要注意存儲路徑的設置?:

獲取樣例文件?
# 進入工作目錄
cd emp-single-end-sequences/# barcode文件
wget \-O barcodes.fastq.gz \https://data.qiime2.org/2023.9/tutorials/moving-pictures/emp-single-end-sequences/barcodes.fastq.gz# 序列文件,這里是單端序列
wget \-O sequences.fastq.gz \https://data.qiime2.org/2023.9/tutorials/moving-pictures/emp-single-end-sequences/sequences.fastq.gz
查看要導入的文件格式:
#
zcat barcodes.fastq.gz | head -n 10
# barcode文件內容,
@HWI-EAS440_0386:1:23:17547:1423#0/1
ATGCAGCTCAGT
+
IIIIIIIIIIIH
@HWI-EAS440_0386:1:23:14818:1533#0/1
CCCCTCAGCGGC
+
DDD@D?@B<<+/
@HWI-EAS440_0386:1:23:14401:1629#0/1
GACGAGTCAGTC#
zcat sequences.fastq.gz | head -n 10# sequences文件內容
@HWI-EAS440_0386:1:23:17547:1423#0/1
TACGNAGGATCCGAGCGTTATCCGGATTTATTGGGTTTAAAGGGAGCGTAGATGGATGTTTAAGTCAGTTGTGAAAGTTTGCGGCTCAACCGTAAAATTGCAGTTGATACTGGATATCTTGAGTGCAGTTGAGGCAGGGGGGGATTGGTGTG
+
IIIE)EEEEEEEEGFIIGIIIHIHHGIIIGIIHHHGIIHGHEGDGIFIGEHGIHHGHHGHHGGHEEGHEGGEHEBBHBBEEDCEDDD>B?BE@@B>@@@@@CB@ABA@@?@@=>?08;3=;==8:5;@6?######################
@HWI-EAS440_0386:1:23:14818:1533#0/1
CCCCNCAGCGGCAAAAATTAAAATTTTTACCGCTTCGGCGTTATAGCCTCACACTCAATCTTTTATCACGAAGTCATGATTGAATCGCGAGTGGTCGGCAGATTGCGATAAACGGGCACATTAAATTTAAACTGATGATTCCACTGCAACAA
+
64<2$24;1)/:*B<?BBDDBBD<>BDD############################################################################################################################
@HWI-EAS440_0386:1:23:14401:1629#0/1
TACGNAGGATCCGAGCGTTATCCGGATTTATTGGGTTTAAAGGGAGCGTAGGCGGACGCTTAAGTCAGTTGTGAAAGTTTGCGGCTCAACCGTAAAATTGCAGTTGATACTGGGTGTCTTGAGTACAGTAGAGGCAGGGGGGGGGTTGGGGG## 激活 qiime2環境:## 導入數據
qiime tools import \--type EMPSingleEndSequences \--input-path ./ \--output-path emp-single-end-sequences.qza#### qiime tools import --show-importable-types 這個命令已經廢棄了,大家注意在新版里沒法用qiime tools list-types
Usage: qiime tools list-types [OPTIONS] [QUERIES]...List the available semantic types.Options:--strict Show only exact matches for the type argument(s).--tsv Print as machine readable tab-separated values.--help Show this message and exit.
雙端序列導入(序列文件與barcode文件各自獨立)

獲取樣例文件??
cd emp-paired-end-sequences
# 下載正向序列
wget \-O forward.fastq.gz \https://data.qiime2.org/2023.9/tutorials/atacama-soils/1p/forward.fastq.gz#下載反向序列
wget \-O reverse.fastq.gz \https://data.qiime2.org/2023.9/tutorials/atacama-soils/1p/reverse.fastq.gz# 下載barcodes文件
wget \-O barcodes.fastq.gz \https://data.qiime2.org/2023.9/tutorials/atacama-soils/1p/barcodes.fastq.gz# 查看輸入文件格式
# 正向序列文件內容
zcat forward.fastq.gz | head -n 10
@M00176:65:000000000-A41FR:1:1101:14282:1412 1:N:0:0
NACGTAGGGTGCAAGCGTTAATCGGAATTACNGGNNNTAAAGCGTGCNNAGGCNNNNNNNNNNNANNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
+
#>>>AAF@ACAA4BGCEEECGGHGGEFCFBG#BA###BABAEFGEEE##BBAA###########B######################################################################################
@M00176:65:000000000-A41FR:1:1101:16939:1420 1:N:0:0
NACGTAGGGGGCAAGCGTTGTCCGGAATCATTGGNNGTAAAGAGCGTGNAGGCNNNNNGNNANNTNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
+
#>>ABAF@ABBBGGGGEEFGGGHGGFFGFHHHHH##BBEFDHHCGGFG#ABFF#####B##B##B######################################################################################
@M00176:65:000000000-A41FR:1:1101:14746:1560 1:N:0:0
TACGTAGGGAGCTAGCGTTGTCCGGAATCATTGGGCGTAAAGCGCGCGTAGGCGGCCAGATAAGTCCGGTGTAAAAGCCACAGGCTNNNNNNNNNNNNNNNNCNGGANNNNNNNNNNNNNNNNNNNNNNANNNNNNNNNNNNANNNNNGGN# 反向序列文件內容
zcat reverse.fastq.gz | head -n 10
@M00176:65:000000000-A41FR:1:1101:14282:1412 2:N:0:0
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCGGAATNCCATNCCNCTCTGCNNNNNNNNNNNNNNNNNNNNNNNNN
+
#########################################################################################################<</?F/#/</?#?<#<???/<#########################
@M00176:65:000000000-A41FR:1:1101:16939:1420 2:N:0:0
NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCNCNNTNCNNNNNNNNNNNGGGAATTCCACNCGCCTCTTCNNNNNNNNNNNNNNNNNNNNNNNNN
+
######################################################################################?#?##?#?###########<??/?FC>FBF#</??FAGHG#########################
@M00176:65:000000000-A41FR:1:1101:14746:1560 2:N:0:0
NNNNTTTCGTNGATNNNNNNNCNGNNNNNNNNNNNNNNNNNNNTNNNNNNNTACCNNNNNNNNTGGGCCTCTGTGCGCATACCGTTCGAGAGCGCCGGCCACGGCATTCGTGTTGCTCCTCTAAGCTGTGCGTGTTCACTGTACAACCNNN# barcode文件內容
zcat barcodes.fastq.gz | head -n 10
@M00176:65:000000000-A41FR:1:1101:14282:1412 1:N:0:0
NNNNNNNNNNNN
+
############
@M00176:65:000000000-A41FR:1:1101:16939:1420 1:N:0:0
NNNNNNNNNNNN
+
############
@M00176:65:000000000-A41FR:1:1101:14746:1560 1:N:0:0
NNNNNNNNNNNN
數據導入:
cd emp-paired-end-sequencesqiime tools import \--type EMPPairedEndSequences \--input-path ./ \--output-path emp-paired-end-sequences.qza
帶barcodes的單端序列文件導入

獲取樣例文件?
#
cd muxed-se-barcode-in-seq/
wget \-O sequences.fastq.gz \https://data.qiime2.org/2023.9/tutorials/importing/muxed-se-barcode-in-seq.fastq.gz# 查看輸入文件格式:
zcat sequences.fastq.gz | head -n 20
@M00899:113:000000000-A5K20:1:1101:18850:2539 1:N:0:2
GCTACGGGGGGCAGCAGTGGGGAATATTGCACAATGGGCGAAAGCCTGATGCAGCAACGCCGCGTGAACGATGAAGGTCTTCGGATCGTAAAGTTCTGTTGCAGGGGAAGATAATGACGGTACCCTGTGAGGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCTAGCGTTATCCGGATTTACTGGGCGTAAAGGGTGCGTAGGTGGTCCTTCAAGTCGGTGGTTAAAGGCTAAGGCTCAACCGTAGTAAGCCGCCGAAACTGGAGGACTTGAGTGAAGGAGAGG
+
-8ABCC>=>5811884:<:99=?@EECFFGDFADECFFFEEDDEFEDEDFFFEEFCCBCF>CCB3CFF:BBFFFCCD,8@9C@C:+5@@:A@C<FDCFBEG>FFFDGCCEC?FGGGGGGGGGCFGGFCFGGGGGGGGGGEG7CFFGFFFGGGFG?FACE;:8CCCCEEF9<F@FFEGGC**/:3:2CC@:C;C81;C9<?>FF8C758CGGG2:7DC>EECEFE9+27CF492/8B7>D)7@F=FFCFF*9F52<2,289<0:44AB<49(3<>F51).69D?D34*44:4<5<B?::086
@M00899:113:000000000-A5K20:1:1101:25454:3578 1:N:0:2
CCTACGGGAGGCAGCAGTGAGGAATATTGGTCAATGGGCGAGAGCCTGAACCAGCCAAGTAGCGTGCAGGATGACGGCCCTATGGGTTGTAAACTGCTTTTGTATGGGGATAAAGTCAGTCACGTGTGATTGTTTGCAGGTACCATACGAATAAGGACCGGCTAATTCCGTGCCAGCAGCCGCGGTAATACGGAAGGTCCGGGCGTTATCCGGATTTATTGGGTTTAAAGGGAGCGTAGGCTGGAGATTAAGTGTGTTGTGAAATGTAGACGCTCAACGTCTGACTTGCAGCGCATACTGG
+
8ACCCGD@AA=18=======;CEFGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGFGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGEGGGGGGFFF?FGGGGGGGGEGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGCGFGGGFFFGGGGGGEFGGGGGGGGGGGGCGEGGDGGGGGGGGGG=CGGCECDGGFGGGGGGGGFGGGF>C>BFFEGFFFFF:FGGF=6=6:AFBGFFFFFFA9A<AFB?@0)>C:0<CF?C46FAD<??90;::?DA>
@M00899:113:000000000-A5K20:1:1101:25177:3605 1:N:0:2
CCTACGGGAGGCAGCAGTGAGGAATATTGGTCAATGGACGGAAGTCTGAACCAGCCAAGTAGCGTGCAGGATGACGGCCCTATGGGTTGTAAACTGCTTTTGTATGGGGATAAAGTTAGGGACGTGTCCCTATTTGCAGGTACCATACGAATAAGGACCGGCTAATTCCGTGCCAGCAGCCGCGGTAATACGGAAGGTCCAGGCGTTATCCGGATTTATTGGGTTTAAAGGGAGCGTAGGCTGGAGATTAAGTGTGTTGTGAAATGTAGACGCTCAACGTCTGAATTGCAGCGCATACTGG
+
88BCCEDAD9018======;;CCFGGGGFGGGFGGGGGGGGGGGGGGGGGGGGGGGFGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGGFGGGGGGGGGGGGGFGGGGGGGGGGGGGGGGEGDGGGGGGGGGFFGGGGGGGGGFGGGFGGGFGGGFFGCGGGGGGFGGFDGGGGGGGGGGGGG5CBFGCGGGGC?FGGGGGGGGGGGDEGDDDGFGGGGGEGGGGGGA39>BFFDDEF4:D5@CE?CFFF>>ABGFFF9<A246<<<<B::DE=<?FGGFG4>F?DF?02211:DAF7### barcode已經在序列中導入文件:
qiime tools import \--type MultiplexedSingleEndBarcodeInSequence \--input-path sequences.fastq.gz \--output-path multiplexed-seqs.qza
當然還有其他可導入文件格式,慢慢看吧,個人覺得有需要的時候查一下就行,不用全部了解:
qiime tools list-formats --importable2、導出數據
官方示例,大家可以下載下來測試一下就行了,不再細講了。
?下載測試:
wget \-O "feature-table.qza" \"https://data.qiime2.org/2023.9/tutorials/exporting/feature-table.qza"wget \-O "unrooted-tree.qza" \"https://data.qiime2.org/2023.9/tutorials/exporting/unrooted-tree.qza"qiime tools export \--input-path feature-table.qza \--output-path exported-feature-table## 導出為biom格式文件
## 參考:https://biom-format.org/documentation/format_versions/biom-2.1.htmlqiime tools export \--input-path unrooted-tree.qza \--output-path exported-tree## 導出為.nwk文件,可在各類軟件再直接出發育樹的圖
head tree.nwk
(((New.CleanUp.ReferenceOTU1480:0.11995,(New.CleanUp.ReferenceOTU202:0.04479,New.CleanUp.ReferenceOTU432:0.0049)0.769:0.04661)1:0.26705,((New.CleanUp.ReferenceOTU1150:0.00016,(New.CleanUp.ReferenceOTU782:0.04264,(New.CleanUp.ReferenceOTU643:0.10438,(((New.CleanUp.ReferenceOTU1014:0.01521,New.CleanUp.ReferenceOTU270:0.02738)0.879:0.02315,(((New.CleanUp.ReferenceOTU1008:0.0378,(New.CleanUp.ReferenceOTU1222:0.01621,(New.CleanUp.ReferenceOTU230:0.01829,(New.CleanUp.ReferenceOTU1047:0.0303,New.CleanUp.ReferenceOTU605:0.00015)0.88:0.01596)0.871:0.01515)0.714:0.01214)0.996:0.06757,(New.CleanUp.ReferenceOTU681:0.00441,(New.CleanUp.ReferenceOTU1485:0.01177,New.CleanUp.ReferenceOTU130:0.05699)0.923:0.02883)0.367:0.02397)0.17:0.00665,(New.CleanUp.ReferenceOTU1330:0.0258,(New.CleanUp.ReferenceOTU903:0.04234,(((New.CleanUp.ReferenceOTU1077:0.00014,(New.CleanUp.ReferenceOTU582:0.0171,New.CleanUp.ReferenceOTU987:0.0226)0.951:0.02372)0.892:0.02206,(New.CleanUp.ReferenceOTU891:0.01038,(((New.CleanUp.ReferenceOTU1066:0.01299,New.CleanUp.ReferenceOTU764:0.02254)0.851:0.01818,(New.CleanUp.ReferenceOTU180:0.02021,New.CleanUp.ReferenceOTU535:0.00014)0.795:0.01084)0.921:0.02441,(New.CleanUp.ReferenceOTU1212:0.00521mkdir extracted-feature-table
qiime tools extract \--input-path feature-table.qza \--output-path extracted-feature-table## 3、樣品元數據處理
這個可能得科學上網,給大家下載了,大家參考下載吧。
https://download.csdn.net/download/zrc_xiaoguo/88616825?spm=1001.2014.3001.5503
樣例文件列表:
sample-metadata數據:

元數據表qiime2轉換命令:
qiime metadata tabulate \--m-input-file sample-metadata.tsv \--o-visualization tabulated-sample-metadata.qzvqiime metadata tabulate \--m-input-file faith_pd_vector.qza \--o-visualization tabulated-faith-pd-metadata.qzvqiime metadata tabulate \--m-input-file sample-metadata.tsv \--m-input-file faith_pd_vector.qza \--o-visualization tabulated-combined-metadata.qzvqiime emperor plot \--i-pcoa unweighted_unifrac_pcoa_results.qza \--m-metadata-file sample-metadata.tsv \--m-metadata-file faith_pd_vector.qza \--o-visualization unweighted-unifrac-emperor-with-alpha.qzv轉換后數據qzv查看
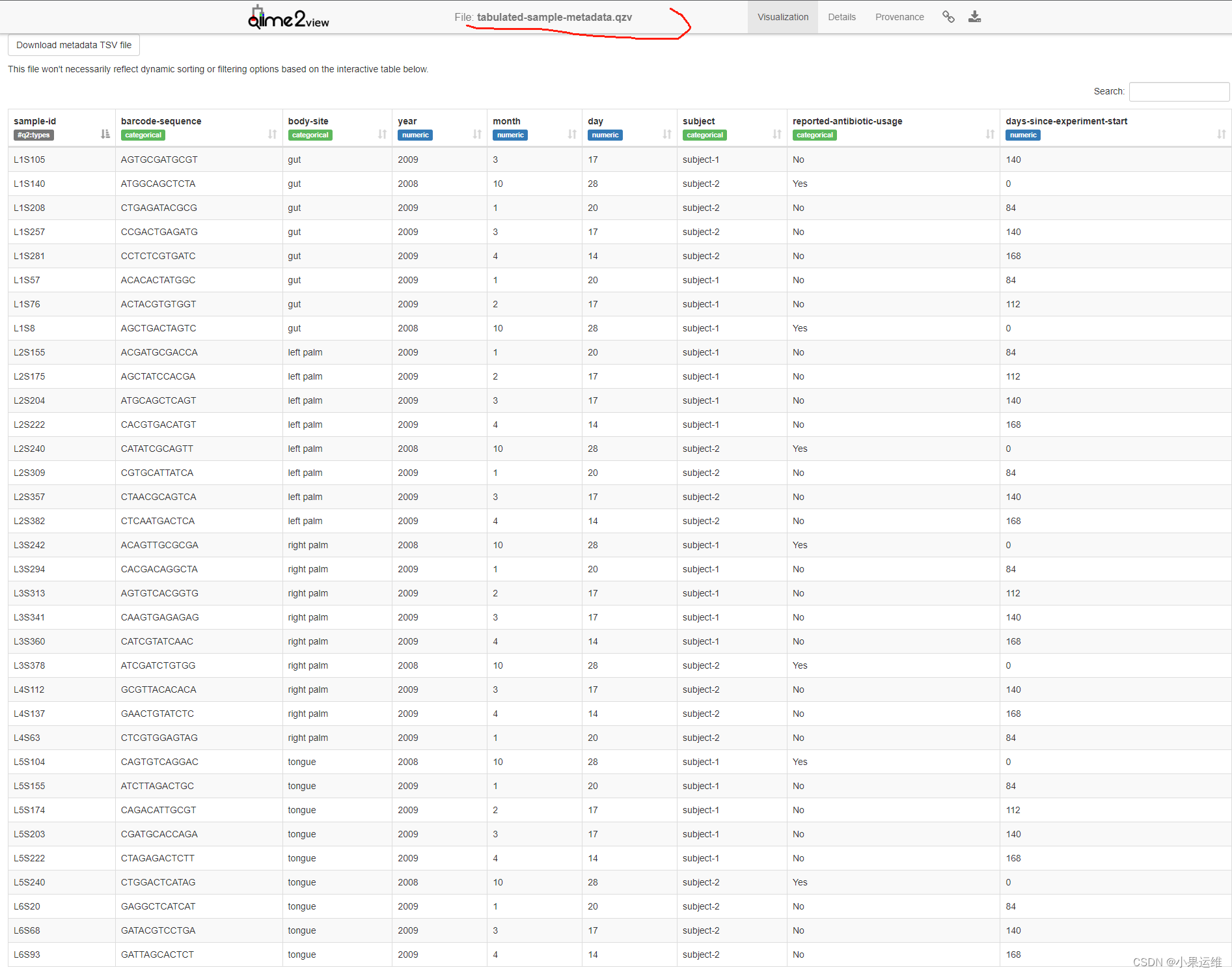
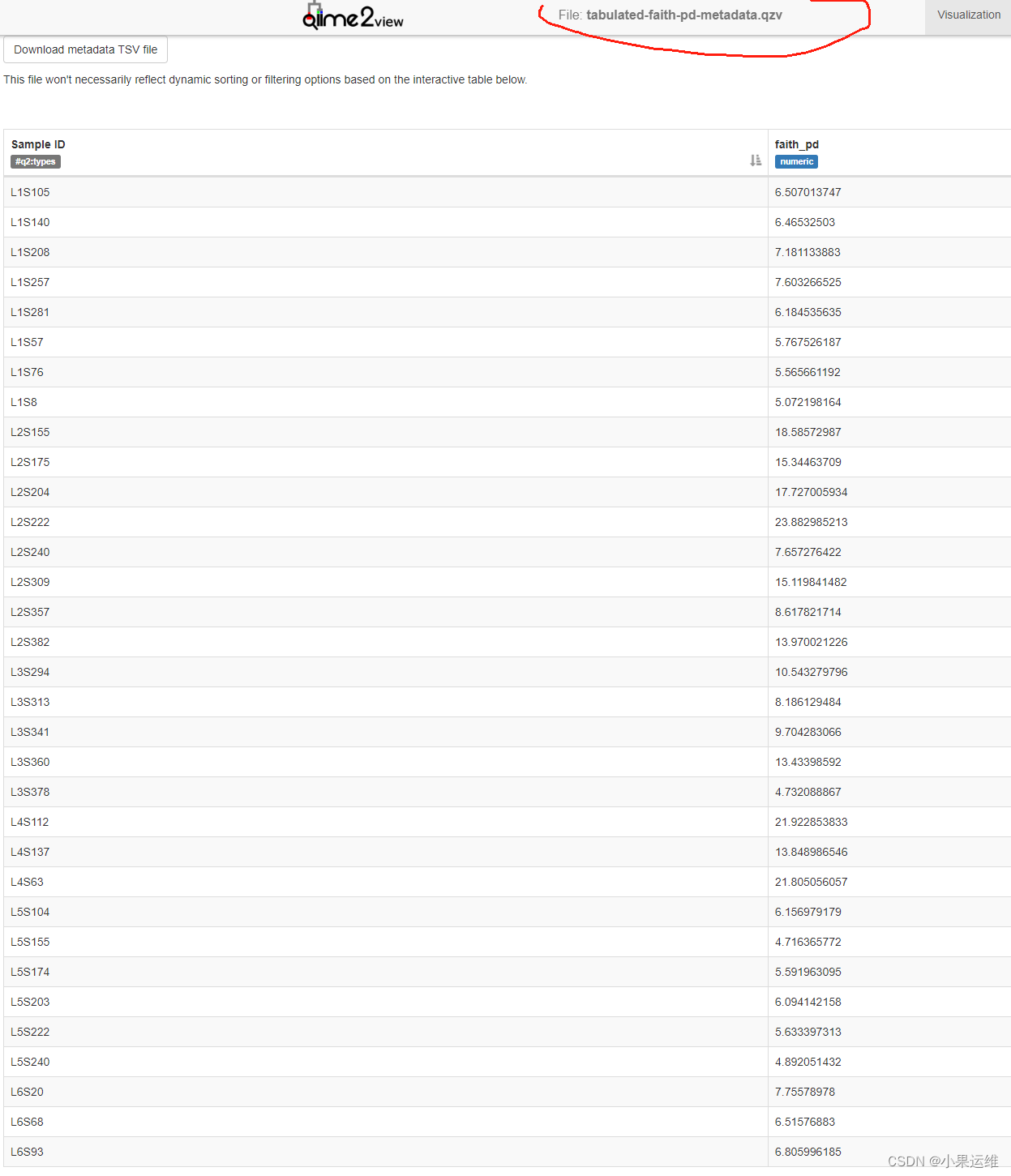
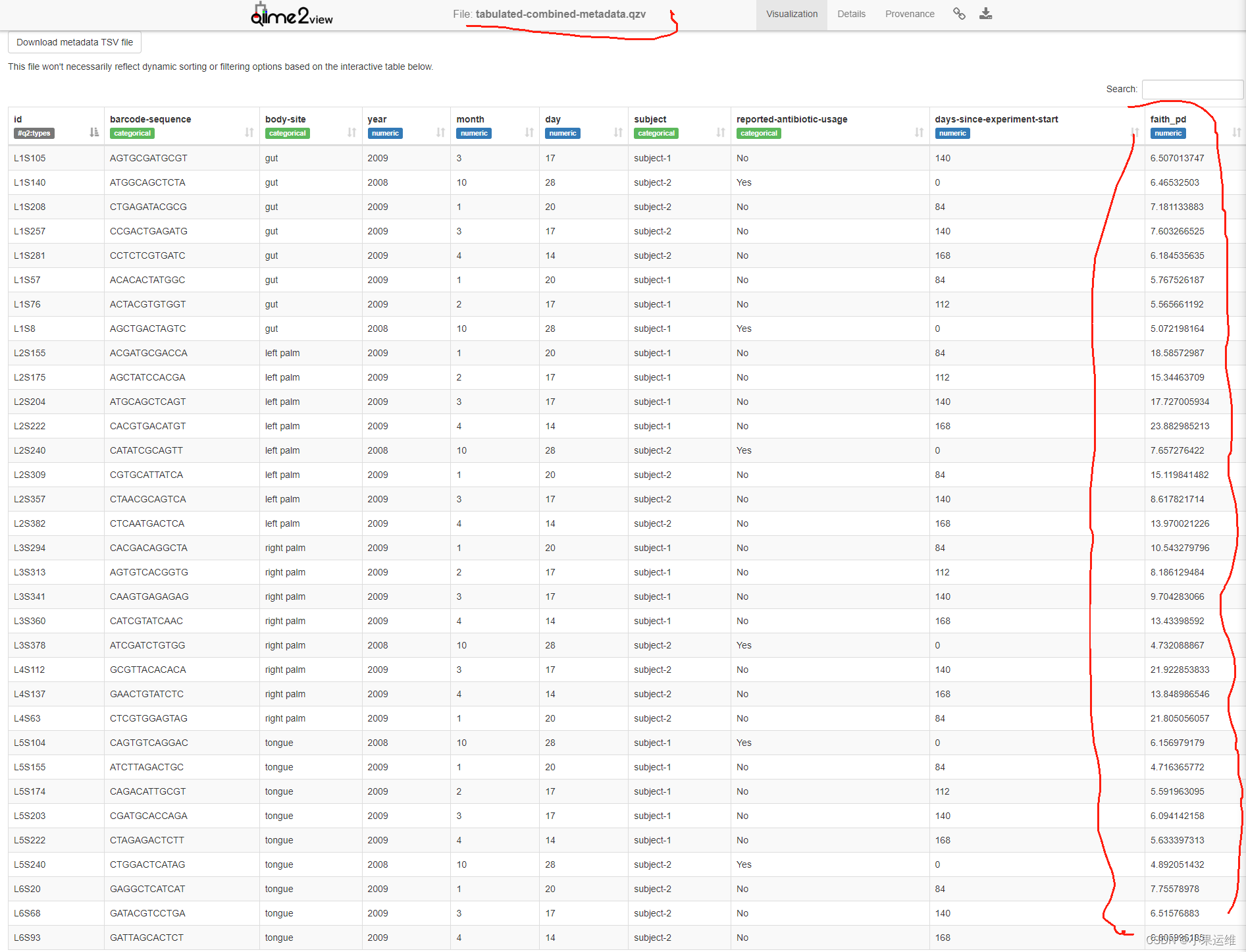
4、數據過濾
Filtering data — QIIME 2 2023.9.2 documentation
特征表過濾Filtering feature tables
基于總出現頻率過濾,去掉低豐度序列,或篩選一定豐度的序列,比如說將頻率低于1500的序列去掉,包括兩個參數,最高和最低頻率
The?--p-min-frequency?and?--p-max-frequency?can be combined to filter based on lower and upper limits of total frequency
這里只給了一個最低頻率的限制。
qiime feature-table filter-samples \--i-table table.qza \--p-min-frequency 1500 \--o-filtered-table sample-frequency-filtered-table.qza基于出現的偶然性的過濾???Contingency-based filtering,也就是最少在多少個樣品中都需要出現,通量有最高和最低頻率設定:?--p-min-features?and?--p-min-samples。
qiime feature-table filter-features \--i-table table.qza \--p-min-samples 2 \--o-filtered-table sample-contingency-filtered-table.qza?字段篩選Identifier-based filtering,?也就是保留哪些特征數據內容。
qiime feature-table filter-samples \--i-table table.qza \--m-metadata-file samples-to-keep.tsv \--o-filtered-table id-filtered-table.qza同樣還有很多其他的篩選方式:
Metadata-based filtering
Taxonomy-based filtering of tables and sequences
序列過濾Filtering sequences
比如說這里根據原序列文件和分類文件提取mitochondria,chloroplast這兩個門水平的序列文件。
qiime taxa filter-seqs \--i-sequences sequences.qza \--i-taxonomy taxonomy.qza \--p-include p__ \--p-exclude mitochondria,chloroplast \--o-filtered-sequences sequences-with-phyla-no-mitochondria-no-chloroplast.qzaFiltering distance matrices
qiime diversity filter-distance-matrix \--i-distance-matrix distance-matrix.qza \--m-metadata-file samples-to-keep.tsv \--o-filtered-distance-matrix identifier-filtered-distance-matrix.qzaqiime diversity filter-distance-matrix \--i-distance-matrix distance-matrix.qza \--m-metadata-file sample-metadata.tsv \--p-where "[subject]='subject-2'" \--o-filtered-distance-matrix subject-2-filtered-distance-matrix.qza5、重要參考數據庫
這個不多說了吧,大家按使用需求來下載,后面逐步會用到:
Data resources — QIIME 2 2023.9.2 documentation
?6、各個插件模塊使用
不熟悉的先查看help信息和官網介紹:
-
q2-alignment
q2-alignment 是 QIIME 2 生態系統中的一個插件,用于對生物信息學序列數據進行比對和序列比較的工具。它可以用于將不同樣本中的序列進行比對,分析它們之間的相似性和差異性。q2-alignment 提供了一些常用的比對算法和工具,使用戶能夠對序列進行比對、生成比對結果和后續的分析。
以下是 q2-alignment 插件的一些主要功能和使用方法:
功能和用途:
- 序列比對:對DNA、RNA或蛋白質序列進行比對。
- 多序列比對:能夠處理多個序列文件,比對它們之間的相似性。
- 生成比對結果:產生比對結果,以便后續分析或可視化。
- 支持多種比對算法:包括常用的比對算法,如BLAST、MUSCLE、MAFFT 等。
qiime alignment --help
Usage: qiime alignment [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin provides support for generating andmanipulating sequence alignments.Plugin website: https://github.com/qiime2/q2-alignmentGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:mafft De novo multiple sequence alignment with MAFFTmafft-add Add sequences to multiple sequence alignment with MAFFT.mask Positional conservation and gap filtering.
# 運行比對(示例使用 MAFFT)
qiime alignment mafft \--i-sequences sequences.qza \--o-alignment aligned_sequences.qza-
q2-composition
用于進行組成分析,尤其是在處理微生物組數據中的相對豐度數據時非常有用。該插件可以幫助用戶探索和比較微生物組中微生物群落的組成。
qiime composition --help
Usage: qiime composition [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin supports methods for compositional dataanalysis.Plugin website: https://github.com/qiime2/q2-compositionGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:add-pseudocount Add pseudocount to table.ancom Apply ANCOM to identify features that differ in abundance.ancombc Analysis of Composition of Microbiomes with Bias Correctionda-barplot Differential abundance bar plotstabulate View tabular output from ANCOM-BC.
# 導入數據
qiime composition add-pseudocount \--i-table table.qza \--o-composition-table composition.qza#數據轉換
qiime composition ilr-transform \--i-table composition.qza \--o-transformed-table ilr_composition.qza#可視化分析結果
qiime composition pcoa \--i-table ilr_composition.qza \--o-pcoa ilr_composition_pcoa.qzaqiime emperor plot \--i-pcoa ilr_composition_pcoa.qza \--m-metadata-file metadata.txt \--o-visualization ilr_composition_emperor.qzv
?q2-cutadapt
q2-cutadapt插件是用于對DNA序列數據進行預處理和過濾的工具。它基于Cutadapt軟件,允許用戶對Illumina測序數據進行裁剪(trimming)、過濾(filtering)和修剪(adapter removal)等操作,以消除低質量序列、去除適配器、修剪序列末端等。這有助于提高序列數據的質量,為后續的分析準備干凈、高質量的數據。
#
qiime cutadapt --help
Usage: qiime cutadapt [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin uses cutadapt to work with adapters (e.g.barcodes, primers) in sequence data.Plugin website: https://github.com/qiime2/q2-cutadaptGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:demux-paired Demultiplex paired-end sequence data with barcodes in-sequence.demux-single Demultiplex single-end sequence data with barcodes in-sequence.trim-paired Find and remove adapters in demultiplexed paired-endsequences.trim-single Find and remove adapters in demultiplexed single-endsequences.qiime cutadapt trim-paired \--i-demultiplexed-sequences demux.qza \--p-cores 8 \--p-front-f CCTACGGGNGGCWGCAG \--p-front-r GACTACHVGGGTATCTAATCC \--p-discard-untrimmed \--o-trimmed-sequences demux_trimmed.qza參數解釋:
--i-demultiplexed-sequences: 輸入的序列文件(需提前導入到QIIME 2)。
--p-cores: 并行處理的CPU核心數量。
--p-front-f 和 --p-front-r: 正向和反向引物序列。
--p-discard-untrimmed: 丟棄未被修剪的序列。
--o-trimmed-sequences: 輸出修剪后的序列文件。?
-
q2-dada2
q2-dada2是QIIME 2的插件之一,基于DADA2算法,用于去噪和分析Illumina測序生成的16S rRNA數據。
qiime dada2 --help
Usage: qiime dada2 [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin wraps DADA2 and supports sequence qualitycontrol for single-end and paired-end reads using the DADA2 R library.Plugin website: http://benjjneb.github.io/dada2/Getting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:denoise-ccs Denoise and dereplicate single-end Pacbio CCSdenoise-paired Denoise and dereplicate paired-end sequencesdenoise-pyro Denoise and dereplicate single-end pyrosequencesdenoise-single Denoise and dereplicate single-end sequences
qiime dada2 denoise-single \--i-demultiplexed-seqs input-demux.qza \--p-trim-left 0 \--p-trunc-len 120 \--o-representative-sequences rep-seqs-dada2.qza \--o-table table-dada2.qza \--o-denoising-stats stats-dada2.qza?參數說明:
--i-demultiplexed-seqs:輸入的demultiplexed序列文件。--p-trim-left:要去除的序列的前部分堿基數量。--p-trunc-len:截斷序列的長度。--o-representative-sequences:輸出的代表序列文件。--o-table:生成的特征表文件。--o-denoising-stats:生成的去噪統計文件。
-
q2-deblur
q2-deblur插件通過識別和去除16S rRNA基因測序數據中的測序錯誤和噪聲,以生成高質量的序列數據。其主要步驟包括:
- 生成特征表(Feature table): 從原始的FASTQ格式文件中導入數據,創建特征表。
- 質量過濾: 過濾低質量序列,去除低質量序列讀數。
- 去噪處理: 使用Deblur算法去除測序錯誤和噪聲,生成高質量的特征序列。
- 生成結果: 輸出一個經過去噪處理和質量過濾的特征表和序列文件。
qiime deblur --help
Usage: qiime deblur [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin wraps the Deblur software for performingsequence quality control.Plugin website: https://github.com/biocore/deblurGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:denoise-16S Deblur sequences using a 16S positive filter.denoise-other Deblur sequences using a user-specified positive filter.visualize-stats Visualize Deblur stats per sample.
qiime deblur denoise-16S \--i-demultiplexed-seqs your_qza_file.qza \--p-trim-length 250 \ # 設置序列截斷長度--o-representative-sequences rep_seqs.qza \--o-table table.qza \--p-sample-stats \ # 如果需要生成樣本統計信息--o-stats deblur_stats.qza
?結果數據導出
# 結果數據導出
qiime tools export --input-path rep_seqs.qza --output-path exported_rep_seqs
qiime tools export --input-path table.qza --output-path exported_table
qiime tools export --input-path deblur_stats.qza --output-path exported_stats
-
q2-demux
q2-demux 插件用于處理 DNA 或 RNA 測序數據的樣本數據解復用(demultiplexing)和質量控制。這個插件允許用戶根據樣本的不同 DNA 或 RNA 序列標簽(barcode 或者 Illumina 測序的 index)將混合測序數據集拆分成單獨的樣本。下面是 q2-demux 插件的基本介紹和使用步驟:
q2-demux 插件功能:
-
數據解復用(Demultiplexing):
- 將混合測序數據根據每個樣本的唯一標識(barcode 或 index)分割成單獨的樣本序列文件。
-
質量控制:
- 提供了檢查序列數據質量的功能,允許用戶查看樣本數據的質量分數并進行必要的處理,比如修剪或過濾低質量序列。
qiime demux --help
Usage: qiime demux [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin supports demultiplexing of single-end andpaired-end sequence reads and visualization of sequence quality information.Plugin website: https://github.com/qiime2/q2-demuxGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:emp-paired Demultiplex paired-end sequence data generatedwith the EMP protocol.emp-single Demultiplex sequence data generated with the EMPprotocol.filter-samples Filter samples out of demultiplexed data.partition-samples-paired Split demultiplexed sequence data into partitions.partition-samples-single Split demultiplexed sequence data into partitions.subsample-paired Subsample paired-end sequences withoutreplacement.subsample-single Subsample single-end sequences withoutreplacement.summarize Summarize counts per sample.tabulate-read-counts Tabulate counts per sample
# 執行去重復
qiime demux emp-paired \--i-seqs demux.qza \--m-barcodes-file sample-metadata.tsv \--m-barcodes-column BarcodeSequence \--o-per-sample-sequences demux-paired-end.qza \--o-error-correction-details demux-details.qza## 查看結果
qiime demux summarize \--i-data demux-paired-end.qza \--o-visualization demux-summary.qzv
?
-
q2-diversity
q2-diversity是其中一個用于計算和分析生物多樣性的插件。它可以幫助你評估樣本群落的多樣性和差異性。
qiime diversity --help
Usage: qiime diversity [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin supports metrics for calculating andexploring community alpha and beta diversity through statistics andvisualizations in the context of sample metadata.Plugin website: https://github.com/qiime2/q2-diversityGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:adonis adonis PERMANOVA test for beta group significancealpha Alpha diversityalpha-correlation Alpha diversity correlationalpha-group-significance Alpha diversity comparisonsalpha-phylogenetic Alpha diversity (phylogenetic)alpha-rarefaction Alpha rarefaction curvesbeta Beta diversitybeta-correlation Beta diversity correlationbeta-group-significance Beta diversity group significancebeta-phylogenetic Beta diversity (phylogenetic)beta-rarefaction Beta diversity rarefactionbioenv bioenvcore-metrics Core diversity metrics (non-phylogenetic)core-metrics-phylogenetic Core diversity metrics (phylogenetic and non-phylogenetic)filter-distance-matrix Filter samples from a distance matrix.mantel Apply the Mantel test to two distance matricespartial-procrustes Partial Procrustespcoa Principal Coordinate Analysispcoa-biplot Principal Coordinate Analysis Biplotprocrustes-analysis Procrustes Analysistsne t-distributed stochastic neighbor embeddingumap Uniform Manifold Approximation and Projection
# Alpha多樣性(樣本內部多樣性)
# 計算Shannon指數:
qiime diversity alpha \--i-table your_feature_table.qza \--p-metric shannon \--o-alpha-diversity shannon_vector.qza#Beta多樣性(樣本間多樣性)
# 計算Bray-Curtis距離:qiime diversity beta \--i-table your_feature_table.qza \--p-metric braycurtis \--o-distance-matrix braycurtis_distance_matrix.qza# 可視化和統計分析
qiime diversity alpha-group-significance \--i-alpha-diversity shannon_vector.qza \--m-metadata-file your_sample_metadata.txt \--o-visualization shannon_group_significance.qzv
qiime diversity beta-group-significance \--i-distance-matrix braycurtis_distance_matrix.qza \--m-metadata-file your_sample_metadata.txt \--o-visualization braycurtis_group_significance.qzv-
q2-diversity-lib
q2-diversity-lib 簡介
q2-diversity-lib是QIIME 2的一個插件,用于計算多樣性指數和樣本間的差異。它基于多種生物多樣性指標來評估微生物群落的多樣性,并允許用戶進行統計比較和可視化。
功能特性
- 計算多樣性指數:支持計算多種多樣性指數,如Shannon、Simpson、Chao1等,可以幫助衡量群落內物種的多樣性和豐富度。
- Beta多樣性計算:計算不同樣本之間的差異和相似性,例如Bray-Curtis、Jaccard、Unweighted UniFrac、Weighted UniFrac等距離指標。
- Beta多樣性可視化:生成多樣性分析的可視化圖表,如PCoA(Principal Coordinates Analysis)圖表,展示樣本之間的差異。
- 組間比較:支持組間多樣性比較,通過PERMANOVA(Permutational Multivariate Analysis of Variance)等方法進行樣本群落的差異性分析。
qiime diversity-lib --help
Usage: qiime diversity-lib [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin computes individual metrics for communityalpha and beta diversity.Plugin website: https://github.com/qiime2/q2-diversity-libGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:alpha-passthrough Alpha Passthrough (non-phylogenetic)beta-passthrough Beta Passthrough (non-phylogenetic)beta-phylogenetic-meta-passthroughBeta Phylogenetic Meta Passthroughbeta-phylogenetic-passthrough Beta Phylogenetic Passthroughbray-curtis Bray-Curtis Dissimilarityfaith-pd Faith's Phylogenetic Diversityjaccard Jaccard Distanceobserved-features Observed Featurespielou-evenness Pielou's Evennessshannon-entropy Shannon's Entropyunweighted-unifrac Unweighted Unifracweighted-unifrac Weighted Unifrac
計算使用?
#計算Alpha多樣性(多樣性指數)
qiime diversity alpha \--i-table table.qza \--p-metric shannon \--o-alpha-diversity shannon_alpha.qza#計算Beta多樣性距離
qiime diversity beta \--i-table table.qza \--p-metric braycurtis \--o-distance-matrix braycurtis_distance.qza#可視化Beta多樣性分析結果(例如PCoA)
qiime diversity pcoa \--i-distance-matrix braycurtis_distance.qza \--o-pcoa braycurtis_pcoa.qza
qiime emperor plot \--i-pcoa braycurtis_pcoa.qza \--m-metadata-file sample-metadata.tsv \--o-visualization braycurtis_emperor.qzv#進行組間多樣性比較(以PERMANOVA為例)
qiime diversity beta-group-significance \--i-distance-matrix braycurtis_distance.qza \--m-metadata-file sample-metadata.tsv \--o-visualization braycurtis_permanova.qzv \--p-method permanova
-
q2-emperor
qiime emperor --help
Usage: qiime emperor [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin wraps Emperor and supports interactivevisualization of ordination plots.Plugin website: http://emperor.microbio.meGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:biplot Visualize and Interact with Principal Coordinates AnalysisBiplotplot Visualize and Interact with Principal Coordinates AnalysisPlotsprocrustes-plot Visualize and Interact with a procrustes plot
-
q2-feature-classifier
q2-feature-classifier 是 QIIME 2 中的一個插件,用于對16S rRNA或ITS等序列數據進行分類和注釋。它主要用于將序列分類為特定的分類單元,比如對OTUs(操作分類單元)或者物種進行分類。
qiime feature-classifier --help
Usage: qiime feature-classifier [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin supports taxonomic classification offeatures using a variety of methods, including Naive Bayes, vsearch, andBLAST+.Plugin website: https://github.com/qiime2/q2-feature-classifierGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:blast BLAST+ local alignment search.classify-consensus-blast BLAST+ consensus taxonomy classifierclassify-consensus-vsearch VSEARCH-based consensus taxonomy classifierclassify-hybrid-vsearch-sklearnALPHA Hybrid classifier: VSEARCH exact match+ sklearn classifierclassify-sklearn Pre-fitted sklearn-based taxonomy classifierextract-reads Extract reads from reference sequences.find-consensus-annotation Find consensus among multiple annotations.fit-classifier-naive-bayes Train the naive_bayes classifierfit-classifier-sklearn Train an almost arbitrary scikit-learnclassifiermakeblastdb Make BLAST database.vsearch-global VSEARCH global alignment search
?
#訓練一個 SILVA 數據庫上的分類器可以使用如下命令
qiime feature-classifier fit-classifier-naive-bayes \--i-reference-reads silva-132-99-515-806-nb-classifier.qza \--i-reference-taxonomy silva-132-99-515-806-nb-classifier.qza \--o-classifier classifier.qza# 使用訓練好的分類器對樣本數據進行分類
qiime feature-classifier classify-sklearn \--i-classifier classifier.qza \--i-reads paired-end-demux.qza \--o-classification taxonomy.qza
-
q2-feature-table
q2-feature-table 插件是 QIIME 2 中的一個重要插件,用于處理和操作特征表(feature table)數據,它包括了許多功能,用于對微生物組數據進行分析和可視化。
以下是 q2-feature-table 插件的一些主要功能和使用方法:
-
導入特征表數據: 可以使用該插件將不同格式的特征表數據導入到 QIIME 2 中,如BIOM格式、文本格式、或其他常見格式的特征表。
-
特征表匯總和統計: 可以對特征表進行匯總和統計描述,比如計算每個樣本中的特征數量、每個特征在樣本中的出現頻率等。
-
特征表的過濾和修剪: 提供了多種方法對特征表進行過濾和修剪,如去除低頻特征、去除低豐度特征、保留指定樣本數或特征數等。
-
特征表的轉換和變換: 可以對特征表進行轉換,如轉置、歸一化、對數轉換等,以適應不同類型的分析需求。
-
特征表的合并和拆分: 可以將多個特征表合并為一個,也可以根據樣本元數據信息將特征表拆分為多個子集。
-
特征表的可視化: 支持對特征表進行可視化展示,比如生成特征數量分布圖、繪制熱圖展示特征在樣本中的豐度等。
qiime feature-table --help
Usage: qiime feature-table [OPTIONS] COMMAND [ARGS]...Description: This is a QIIME 2 plugin supporting operations on sample byfeature tables, such as filtering, merging, and transforming tables.Plugin website: https://github.com/qiime2/q2-feature-tableGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:core-features Identify core features in tablefilter-features Filter features from tablefilter-features-conditionally Filter features from a table based onabundance and prevalencefilter-samples Filter samples from tablefilter-seqs Filter features from sequencesgroup Group samples or features by a metadatacolumnheatmap Generate a heatmap representation of afeature tablemerge Combine multiple tablesmerge-seqs Combine collections of feature sequencesmerge-taxa Combine collections of feature taxonomiespresence-absence Convert to presence/absencerarefy Rarefy tablerelative-frequency Convert to relative frequenciesrename-ids Renames sample or feature ids in a tablesplit Split one feature table into manysubsample Subsample tablesummarize Summarize tabletabulate-seqs View sequence associated with each featuretranspose Transpose a feature table.
#特征表的摘要和統計信息
qiime feature-table summarize \--i-table feature-table.qza \--o-visualization feature-table-summary.qzv#特征表的過濾和修剪
qiime feature-table filter-features \--i-table feature-table.qza \--p-min-frequency 10 \--o-filtered-table filtered-feature-table.qzaqiime feature-table filter-samples \--i-table feature-table.qza \--p-min-frequency 500 \--o-filtered-table filtered-sample-table.qza# 特征表的合并和操作
qiime feature-table merge \--i-tables table1.qza \--i-tables table2.qza \--o-merged-table merged-table.qza#計算特征表的β-diversity:
qiime diversity beta \--i-table feature-table.qza \--o-distance-matrix beta-diversity.qza \--p-metric braycurtis#可視化特征表
qiime feature-table summarize \--i-table feature-table.qza \--o-visualization feature-table-summary.qzv#可視化β-diversity距離矩陣
qiime diversity beta-phylogenetic \--i-table feature-table.qza \--i-phylogeny rooted-tree.qza \--o-distance-matrix beta-diversity.qza \--p-metric unweighted_unifracqiime diversity pcoa \--i-distance-matrix beta-diversity.qza \--o-pcoa pcoa-results.qzaqiime emperor plot \--i-pcoa pcoa-results.qza \--m-metadata-file sample-metadata.tsv \--o-visualization emperor.qzv
-
q2-fragment-insertion
q2-fragment-insertion插件是用于將未分配的DNA序列(通常是16S rRNA或18S rRNA序列)嵌入(插入)到預先構建的參考進化樹中的工具。這個插件可以幫助解決一些問題,比如通過將未知序列嵌入到進化樹中,來推斷未知序列的系統發育位置。
qiime fragment-insertion --help
Usage: qiime fragment-insertion [OPTIONS] COMMAND [ARGS]...Description: No description available. See plugin website:https://github.com/qiime2/q2-fragment-insertionPlugin website: https://github.com/qiime2/q2-fragment-insertionGetting user support: https://github.com/qiime2/q2-fragment-insertion/issuesOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:classify-otus-experimental Experimental: Obtain taxonomic lineages, byfinding closest OTU in reference phylogeny.filter-features Filter fragments in tree from table.sepp Insert fragment sequences using SEPP intoreference phylogenies.
qiime fragment-insertion sepp \--i-representative-sequences <代表序列文件.qza> \--i-reference-database <參考數據庫.qza> \--o-tree <輸出進化樹.qza> \--o-placements <輸出插入位置文件.qza>
--i-representative-sequences是代表性序列文件的位置。--i-reference-database是參考數據庫文件的位置。--o-tree指定輸出的進化樹文件。--o-placements指定輸出的插入位置文件。
-
q2-longitudinal
q2-longitudinal 插件是 QIIME 2 中的一個插件,專門用于處理微生物組長期研究的數據。該插件允許用戶對時間序列實驗數據進行分析,以便檢測微生物組隨時間變化的情況,比較不同條件下的變化,以及對這些變化的統計顯著性進行評估。
以下是 q2-longitudinal 插件的一些主要功能和使用命令:
主要功能:
- 時間序列數據可視化:生成時間序列樣本數據的可視化圖表,比如長期研究的變化趨勢、樣本之間的差異等。
- 差異分析:比較不同時間點或不同處理組之間的微生物組成差異。
- Alpha 和 Beta 多樣性分析:評估微生物群落在時間序列中的多樣性和相似性變化。
- 線性混合效應模型:對微生物組數據進行線性模型分析,以研究時間、處理效應和其交互作用對微生物組成的影響。
qiime longitudinal --help
Usage: qiime longitudinal [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin supports methods for analysis of timeseries data, involving either paired sample comparisons or longitudinalstudy designs.Plugin website: https://github.com/qiime2/q2-longitudinalGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:anova ANOVA testfeature-volatility Feature volatility analysisfirst-differences Compute first differences or difference frombaseline between sequential statesfirst-distances Compute first distances or distance from baselinebetween sequential stateslinear-mixed-effects Linear mixed effects modelingmaturity-index Microbial maturity index prediction.nmit Nonparametric microbial interdependence testpairwise-differences Paired difference testing and boxplotspairwise-distances Paired pairwise distance testing and boxplotsplot-feature-volatility Plot longitudinal feature volatility andimportancesvolatility Generate interactive volatility plot
#導入數據
qiime longitudinal feature-table-merge \--i-longitudinal-feature-tables feature-table.qza \--o-merged-table merged-table.qza#線性混合效應模型
qiime longitudinal linear-mixed-effects \--m-metadata-file sample-metadata.tsv \--m-metadata-file timepoints.tsv \--p-metric Shannon_index \--p-group-columns treatment \--p-state-column time \--p-individual-id-column subject \--o-visualization lme-results.qzv#可視化
qiime longitudinal volatility \--i-table merged-table.qza \--m-metadata-file sample-metadata.tsv \--p-state-column time \--p-individual-id-column subject \--o-visualization volatility.qzv#組間差異
qiime longitudinal pairwise-differences \--m-metadata-file sample-metadata.tsv \--m-metadata-file timepoints.tsv \--p-metric Shannon_index \--p-group-column treatment \--p-state-column time \--p-individual-id-column subject \--o-visualization pairwise-differences.qzv#α多樣性圖表
qiime longitudinal maturity-index \--i-alpha-diversity alpha-diversity.qza \--m-metadata-file sample-metadata.tsv \--p-state-column time \--p-individual-id-column subject \--o-visualization maturity-index.qzv
-
q2-metadata
q2-metadata 插件用于處理和操作元數據,元數據是描述樣本信息的數據,比如樣本來源、處理方法、實驗條件等。以下是 q2-metadata 插件的簡要介紹和一些常見的使用命令:
q2-metadata 插件的功能:
- 元數據導入: 將元數據文件導入到 QIIME 2 格式中。
- 元數據可視化: 可視化元數據內容以便更好地理解樣本信息。
- 元數據的處理和編輯: 對元數據進行篩選、編輯和轉換。
- 元數據統計和摘要: 統計和生成關于元數據的摘要信息。
qiime metadata --help
Usage: qiime metadata [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin provides functionality for working with andvisualizing Metadata.Plugin website: https://github.com/qiime2/q2-metadataGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:distance-matrix Create a distance matrix from a numeric Metadata columnmerge Merge metadatashuffle-groups Shuffle values in a categorical sample metadata column.tabulate Interactively explore Metadata in an HTML table
# 1. 元數據導入:
qiime metadata tabulate --m-input-file sample-metadata.tsv --o-visualization sample-metadata.qzv
# 2. 元數據可視化:
qiime metadata tabulate --m-input-file sample-metadata.qza --o-visualization sample-metadata.qzv
# 3. 元數據編輯與處理:
# 刪除列:
qiime metadata tabulate --m-input-file sample-metadata.qza --o-visualization sample-metadata.qzv
# 篩選行(過濾):
qiime metadata tabulate --m-input-file sample-metadata.qza --o-visualization sample-metadata.qzv
# 4. 元數據統計和摘要:
qiime metadata tabulate --m-input-file sample-metadata.qza --o-visualization sample-me-
q2-phylogeny
q2-phylogeny插件則提供了處理生物多樣性分析中的系統發育信息的功能。該插件主要用于構建系統發育樹和處理系統發育樹相關的操作,例如序列的進化樹推斷、進化樹的根節點分配等。
qiime phylogeny --help
Usage: qiime phylogeny [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin supports generating and manipulatingphylogenetic trees.Plugin website: https://github.com/qiime2/q2-phylogenyGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:align-to-tree-mafft-fasttree Build a phylogenetic tree using fasttree andmafft alignmentalign-to-tree-mafft-iqtree Build a phylogenetic tree using iqtree andmafft alignment.align-to-tree-mafft-raxml Build a phylogenetic tree using raxml andmafft alignment.fasttree Construct a phylogenetic tree with FastTree.filter-table Remove features from table if they're notpresent in tree.filter-tree Remove features from tree based on metadataiqtree Construct a phylogenetic tree with IQ-TREE.iqtree-ultrafast-bootstrap Construct a phylogenetic tree with IQ-TREEwith bootstrap supports.midpoint-root Midpoint root an unrooted phylogenetic tree.raxml Construct a phylogenetic tree with RAxML.raxml-rapid-bootstrap Construct a phylogenetic tree with bootstrapsupports using RAxML.robinson-foulds Calculate Robinson-Foulds distance betweenphylogenetic trees.
## 構建系統發育樹:
#使用多序列比對后的結果構建系統發育樹。可以使用q2-phylogeny中的FastTree或RAxML進行樹的構建。以下是使用FastTree的示例命令:
qiime phylogeny fasttree \--i-alignment aligned-sequences.qza \--o-tree tree.qza# 或者使用RAxML進行系統發育樹的構建:
qiime phylogeny raxml \--i-alignment aligned-sequences.qza \--p-substitution-model GTRGAMMA \--o-tree tree.qza \--verbose## 可選操作 - 根節點分配:
# 有時,你可能需要為系統發育樹分配根節點。可以使用q2-phylogeny中的根節點分配插件進行此操作。以下是一個示例命令:
qiime phylogeny midpoint-root \--i-tree tree.qza \--o-rooted-tree rooted-tree.qza?
-
q2-quality-control
q2-quality-control 插件旨在進行序列數據的質量控制和過濾,它可以執行以下任務:
- 對序列數據進行質量評估
- 去除低質量序列
- 截取或修剪序列的部分
- 去除嵌合序列(chimeras)
- 過濾低頻序列
-
q2-quality-filter
q2-quality-filter插件用于對DNA測序數據進行質量控制和過濾,以去除低質量的序列。這有助于提高后續分析的準確性和可靠性。
qiime quality-control --help
Usage: qiime quality-control [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin supports methods for assessing andcontrolling the quality of feature and sequence data.Plugin website: https://github.com/qiime2/q2-quality-controlGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:bowtie2-build Build bowtie2 index from reference sequences.decontam-identify Identify contaminantsdecontam-identify-batches Identify contaminants in Batch Modedecontam-remove Removes contaminantdecontam-score-viz Generate a histogram representation of the scoresevaluate-composition Evaluate expected vs. observed taxonomiccomposition of samplesevaluate-seqs Compare query (observed) vs. reference (expected)sequences.evaluate-taxonomy Evaluate expected vs. observed taxonomicassignmentsexclude-seqs Exclude sequences by alignmentfilter-reads Filter demultiplexed sequences by alignment toreference database.
qiime quality-filter q-score \--i-demux paired-end-demux.qza \--p-min-quality 20 \--o-filtered-sequences demux-filtered.qza \--o-filter-stats demux-filter-stats.qza
-
q2-sample-classifier
而q2-sample-classifier是Qiime 2中的一個插件,用于樣本分類和預測。它可以幫助用戶利用機器學習算法對樣本進行分類,比較不同條件下的微生物組成差異,例如,預測分類數據(如臨床數據)和微生物組成之間的關系。
qiime sample-classifier --help
Usage: qiime sample-classifier [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin supports methods for supervisedclassification and regression of sample metadata, and other supervisedmachine learning methods.Plugin website: https://github.com/qiime2/q2-sample-classifierGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:classify-samples Train and test a cross-validated supervisedlearning classifier.classify-samples-from-dist Run k-nearest-neighbors on a labeled distancematrix.classify-samples-ncv Nested cross-validated supervised learningclassifier.confusion-matrix Make a confusion matrix from sample classifierpredictions.fit-classifier Fit a supervised learning classifier.fit-regressor Fit a supervised learning regressor.heatmap Generate heatmap of important features.metatable Convert (and merge) positive numeric metadata(in)to feature table.predict-classification Use trained classifier to predict target valuesfor new samples.predict-regression Use trained regressor to predict target valuesfor new samples.regress-samples Train and test a cross-validated supervisedlearning regressor.regress-samples-ncv Nested cross-validated supervised learningregressor.scatterplot Make 2D scatterplot and linear regression ofregressor predictions.split-table Split a feature table into training and testingsets.summarize Summarize parameter and feature extractioninformation for a trained estimator.
qiime sample-classifier classify-samples \--i-table feature-table.qza \--m-metadata-file sample-metadata.qza \--m-metadata-column TARGET_COLUMN \--p-test-size 0.2 \--p-random-state 42 \--p-n-estimators 100 \--p-n-jobs 1 \--o-visualization classification-results.qzv
--i-table指定特征表格的位置--m-metadata-file指定樣本元數據的位置--m-metadata-column指定用于分類的目標列--p-test-size設置測試集的比例--p-random-state設置隨機種子以確保結果可重復--p-n-estimators設置分類器使用的估計器數量--p-n-jobs設置用于計算的作業數--o-visualization指定輸出結果的位置
-
q2-taxa
q2-taxa 插件則用于分析和可視化分類學信息,特別是對于已經進行了序列分類(比如16S rRNA)的數據。q2-taxa 允許用戶對物種注釋信息進行處理、匯總和可視化。
qiime taxa --help
Usage: qiime taxa [OPTIONS] COMMAND [ARGS]...Description: This QIIME 2 plugin provides functionality for working with andvisualizing taxonomic annotations of features.Plugin website: https://github.com/qiime2/q2-taxaGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:barplot Visualize taxonomy with an interactive bar plotcollapse Collapse features by their taxonomy at the specified levelfilter-seqs Taxonomy-based feature sequence filter.filter-table Taxonomy-based feature table filter.
# 1. 導入數據
# 使用命令 qiime taxa 可以導入并生成用于分類學分析的數據格式。例如:
qiime taxa import \--input-path taxonomy.tsv \--output-path taxonomy.qza \--type 'FeatureData[Taxonomy]'# 2. 查看物種豐度
# 命令 qiime taxa barplot 可以用于生成物種豐度的柱狀圖,用于可視化各個樣本中不同分類水平的相對豐度。
qiime taxa barplot \--i-table table.qza \--i-taxonomy taxonomy.qza \--m-metadata-file sample-metadata.tsv \--o-visualization taxa-bar-plot.qzv# 3. 物種注釋
# 可通過 qiime taxa classify-sklearn 命令使用機器學習分類器進行序列的物種注釋。以下是一個例子:
qiime taxa classify-sklearn \--i-reads rep-seqs.qza \--i-classifier classifier.qza \--o-classification taxonomy.qza# 4. 特定分類水平的篩選和可視化
# 可以使用 qiime taxa filter-table 和 qiime taxa filter-seqs 命令在特定分類水平對數據進行過濾。然后,可以使用其他命令可視化已過濾的數據。# 5. 物種多樣性分析
使用 qiime taxa collapse 和 qiime taxa rarefy 等命令可以對物種注釋數據進行聚合和稀釋,以進行多樣性分析。-
q2-types
q2-types插件旨在處理和管理不同類型的數據,包括:
- DNA 序列
- RNA 序列
- 蛋白質序列
- 樣本和元數據信息
-
q2-vsearch
?q2-vsearch 插件則是 QIIME 2 中用于序列相似性搜索和聚類的插件之一。它基于 VSEARCH,提供了執行序列相似性搜索、聚類和序列分析的功能。這個基本上與直接使用vesearch直接使用差不多了,只不過這里集成后可以直接與qiime的其他模塊相結合使用。
qiime vsearch --help
Usage: qiime vsearch [OPTIONS] COMMAND [ARGS]...Description: This plugin wraps the vsearch application, and provides methodsfor clustering and dereplicating features and sequences.Plugin website: https://github.com/qiime2/q2-vsearchGetting user support: Please post to the QIIME 2 forum for help with thisplugin: https://forum.qiime2.orgOptions:--version Show the version and exit.--example-data PATH Write example data and exit.--citations Show citations and exit.--help Show this message and exit.Commands:cluster-features-closed-referenceClosed-reference clustering of features.cluster-features-de-novo De novo clustering of features.cluster-features-open-referenceOpen-reference clustering of features.dereplicate-sequences Dereplicate sequences.fastq-stats Fastq stats with vsearch.merge-pairs Merge paired-end reads.uchime-denovo De novo chimera filtering with vsearch.uchime-ref Reference-based chimera filtering with
# 序列相似性搜索
# 使用 q2-vsearch 插件進行序列相似性搜索的命令:
qiime vsearch search-sequences \--i-query query_sequences.qza \--i-reference reference_sequences.qza \--o-results search_results.qza \--o-perc-identity output_identity.qza \--o-failed-sequences failed_sequences.qza
# 此命令將執行一項序列搜索,將查詢序列與參考序列集進行比較,并生成包含搜索結果的文件。# 序列聚類
# 利用 q2-vsearch 進行序列聚類的命令示例:
qiime vsearch cluster-features-de-novo \--i-sequences sequences.qza \--i-table table.qza \--p-perc-identity 0.97 \--o-clustered-table clustered_table.qza \--o-clustered-sequences clustered_sequences.qza \--o-new-reference-sequences new_reference_sequences.qza當然還有分析結果的可視化
在線可視化,拖拽出圖:QIIME 2 View
?






)
信息系統基礎知識)

)




)



)
)