歡迎關注我的CSDN:https://spike.blog.csdn.net/
本文地址:https://spike.blog.csdn.net/article/details/134599076
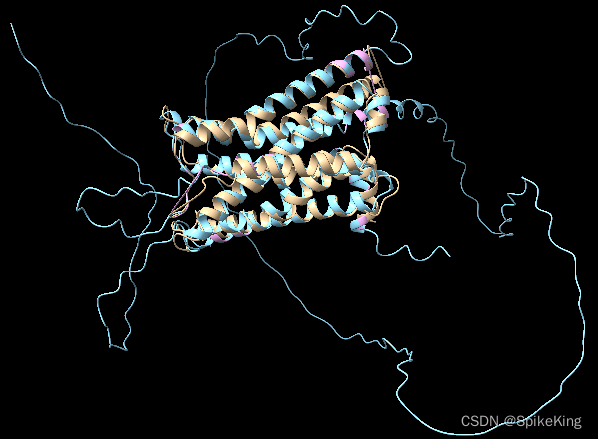
在蛋白質結構預測的過程中,輸入一般是蛋白質序列(長序列),預測出 PDB 三維結構,再和 Ground Truth 的 PDB 進行比較,GT 的 PDB 是實驗測出,比真實的蛋白質序列要短,需要使用算法進行查找。
滿足約束:PDB 結構序列的長度 小于 蛋白質序列的長度,并且是子集關系。
- 黃色是 PDB 官網的結構
- 藍色是預測的全長序列的結構
- 粉色是通過算法,截取的子結構
即:
源碼:
def match_sub_seq(seq_long, seq_short):"""匹配序列子串,返回區間范圍,一般用于長FASTA與短PDBSeq之間的預處理"""def get_seq_max_idx(seq_l, seq_s):"""序列匹配結果,返回連續最大索引"""np = len(seq_l)nf = len(seq_s)res = [0] * npi, j = 0, 0same = 0is_next, next_i = True, 0while i < np:rp = seq_l[i] # pdb 的 殘基rf = seq_s[j] # fasta 的殘基if is_next:next_i = i + 1is_next = Falseif rp == rf:same += 1j += 1else:j = 0same = 0if seq_l[i] == seq_s[j]:same += 1j += 1if i < np:res[i] = max(same, res[i])i += 1if j >= nf:j = 0same = 0# 避免 "AAABCDEFGAB" 與 "AABCDEFG" 情況if rp != rf:i = next_iis_next = Truereturn resnl, ns = len(seq_long), len(seq_short)size = 0gap_list = [] # 區間范圍tmp_seq_short = seq_short# print(f"[Info] seq_long: {seq_long}")# print(f"[Info] seq_short: {seq_short}")prev_idx = 0while size < ns:# print(f"[Info] tmp_seq_short: {tmp_seq_short}")res = get_seq_max_idx(seq_long, tmp_seq_short)max_val = max(res) # 最大索引值max_indices = [i for i, x in enumerate(res) if x == max_val]for j in sorted(max_indices):# print(j, prev_idx)if j <= prev_idx:continueelse:max_idx = jbreaks_idx, e_idx = max_idx-max_val+1, max_idx+1prev_idx = e_idxgap_list.append([s_idx, e_idx])tmp_sub_long = seq_long[s_idx:e_idx]tmp_short = tmp_seq_short[:max_val]assert tmp_sub_long == tmp_shorttmp_seq_short = tmp_seq_short[max_val:]size += max_val# 驗證邏輯f_seq = ""for gap in gap_list:f_seq += seq_long[gap[0]:gap[1]]assert f_seq == seq_shortreturn gap_list
從索引中,提取 PDB:
def extract_pdb_from_gap(pdb_path, output_path, gap_list):"""從殘基的 gap list 提取新的 PDB 文件"""d3to1 = {'CYS': 'C', 'ASP': 'D', 'SER': 'S', 'GLN': 'Q', 'LYS': 'K','ILE': 'I', 'PRO': 'P', 'THR': 'T', 'PHE': 'F', 'ASN': 'N','GLY': 'G', 'HIS': 'H', 'LEU': 'L', 'ARG': 'R', 'TRP': 'W','ALA': 'A', 'VAL': 'V', 'GLU': 'E', 'TYR': 'Y', 'MET': 'M'}d1to3 = invert_dict(d3to1)# chain_idx = 0atom_num_idx = 1res_seq_num_idx = 0pre_res_seq_num = "" # 殘基可能是52Achain_id_list = []out_lines = []line_idx = 0lines = read_file(pdb_path)res_ca_dict = dict()for idx, line in enumerate(lines):# 只處理核心行record_type = str(line[:6].strip()) # 1~6if record_type not in ["ATOM", "HETATM"]:continuerecord_type = "ATOM"line_idx += 1# 重新設置 atom_serial_num# atom_num = int(line[6:11].strip()) # 7~11atom_num = atom_num_idxatom_num_idx += 1# 替換為標準氨基酸residue_name = str(line[17:20].strip()) # 18~20if residue_name not in d3to1_ex:continueif residue_name in d3to1_ex and residue_name not in d3to1.keys():a = d3to1_ex[residue_name]residue_name = d1to3[a]# 不修改鏈名chain_id = str(line[21].strip()) # 22if chain_id not in chain_id_list: # 更換鏈chain_id_list.append(chain_id)pre_res_seq_num = ""# chain_idx += 1# chain_id = chr(ord("A") + chain_idx - 1)# 重新設置 res_seq_numres_seq_num = line[22:27].strip() # 23~26 \ 23~27if pre_res_seq_num != res_seq_num: # 更換殘基pre_res_seq_num = res_seq_numres_seq_num_idx += 1res_ca_dict[res_seq_num_idx] = Falseres_seq_num = res_seq_num_idx# 確保只有一個CAatom_name = str(line[12:16].strip()) # 13~16if atom_name == "CA":if res_ca_dict[res_seq_num]:print(f"[Warning] PDB res {res_seq_num} has more than one CA! ")continueelse:res_ca_dict[res_seq_num] = Truecoordinates_x = str(line[30:38].strip()) # 31~38coordinates_y = str(line[38:46].strip()) # 39~46coordinates_z = str(line[46:54].strip()) # 47~54occupancy = str(line[54:60].strip()) # 55~60temperature_factor = str(line[60:66].strip()) # 61~66element_symbol = str(line[76:78]) # 77~78# 判斷殘基索引是否在 gap_list 中,其余保持不變is_skip = Truefor gap in gap_list:if gap[0] <= res_seq_num-1 < gap[1]:is_skip = Falseif is_skip:continueout_line = "{:<6}{:>5} {:^4} {:<3} {:<1}{:>4} {:>8}{:>8}{:>8}{:>6}{:>6} {:>2}".format(str(record_type), str(atom_num), str(atom_name), str(residue_name),str(chain_id), str(res_seq_num),str(coordinates_x), str(coordinates_y), str(coordinates_z),str(occupancy), str(temperature_factor),str(element_symbol))out_lines.append(out_line)create_empty_file(output_path)write_list_to_file(output_path, out_lines)def truncate_pdb_by_sub_seq(pdb_path, sub_seq, output_path):"""根據子序列提取新的 PDB 文件"""seq_str, n_chains, chain_dict = get_seq_from_pdb(pdb_path)pdb_seq = list(chain_dict.values())[0]try:gap_list = match_sub_seq(pdb_seq, sub_seq)except Exception as e:print(f"[Warning] input sub_seq is not pdb sub seq! {pdb_path}")return output_pathextract_pdb_from_gap(pdb_path, output_path, gap_list)# 驗證邏輯seq_str, n_chains, chain_dict = get_seq_from_pdb(output_path)new_seq = list(chain_dict.values())[0]assert new_seq == sub_seq, print(f"[Error] new_seq: {new_seq}, sub_seq: {sub_seq}")return output_path











】)
 細說 評估指標的交叉驗證)




)

