Title
題目
Hierarchical Vision Transformers for prostate biopsy grading: Towardsbridging the generalization gap
用于前列腺活檢分級的分層視覺 Transformer:邁向彌合泛化差距
01
文獻速遞介紹
前列腺癌是全球男性中第二常見的確診癌癥,也是第五大致命癌癥。病理學家對前列腺活檢樣本進行分級,在確定前列腺癌的侵襲性方面起著關鍵作用,進而指導從主動監測到手術等一系列干預措施。隨著前列腺癌患者數量不斷增加,病理學家的工作壓力日益增大,因此迫切需要借助計算方法輔助日常工作流程。 組織切片的數字化使得大規模數據集的可獲得性不斷提高,這為計算機視覺研究創造了機會,有助于通過深度學習算法為病理學家提供支持和輔助。深度學習徹底改變了計算機視覺的多個領域,在圖像分類、目標檢測和語義分割等任務中取得了前所未有的成功。十年前,卷積神經網絡(CNNs)取得了重大進展(Krizhevsky 等人,2012),而近年來,諸如視覺Transformer(ViT)(Dosovitskiy 等人,2021)等基于注意力機制的模型進一步突破了性能極限。 然而,由于全切片圖像(WSIs)尺寸極大,超出了傳統深度學習硬件的內存容量,計算病理學面臨著一系列獨特的挑戰。因此,人們提出了創新策略來克服這一內存瓶頸。一種主流方法是將這些龐大的圖像分割成更小的補丁。這些補丁通常作為輸入單元,其標簽來自像素級注釋(Ehteshami Bejnordi 等人,2017;Coudray 等人,2018)。但獲取像素級注釋既耗時又不切實際,尤其是在前列腺癌分級等復雜任務中,病理學家只能對他們能識別的部分進行注釋。因此,近年來的研究探索了超越全監督的技術,重點關注更靈活的訓練范式,如弱監督學習。 弱監督學習利用粗粒度(圖像級)信息自動推斷細粒度(補丁級)細節。多實例學習(MIL)近年來在多項計算病理學挑戰中成為一種強大的弱監督方法,并展現出卓越的性能(Hou 等人,2015;Campanella 等人,2019)。通過僅使用切片級標簽就能對全切片圖像進行分析,它避開了對像素級注釋的需求,為風險預測和基因突變檢測等任務提供了便利(Schmauch 等人,2020;Garberis 等人,2022)。 盡管多實例學習取得了成功,但大多數多實例學習方法忽略了補丁之間的空間關系,從而錯失了有價值的上下文信息。為解決這一局限,研究重點轉向開發能夠整合更廣泛上下文的方法(Lerousseau 等人,2021;Pinckaers 等人,2021;Shao 等人,2021)。其中,分層視覺Transformer(H-ViTs)已成為一種很有前景的解決方案,在癌癥亞型分類和生存預測方面取得了最先進的成果(Chen 等人,2022)。 基于這些考慮,我們在前列腺癌分級背景下對分層視覺Transformer進行了全面分析。我們的工作在該領域取得了多項關鍵進展,具體如下: 1. 我們發現,當在與訓練數據來自同一中心的病例上進行測試時,分層視覺Transformer與最先進的前列腺癌分級算法性能相當,同時在更多樣化的臨床場景中表現出更強的泛化能力。 2. 我們證明了針對前列腺的特異性預訓練相比更通用的多器官預訓練具有優勢。 3. 我們系統地比較了兩種分層視覺Transformer變體,并就每種變體更適用的場景提供了具體指導。 4. 我們對序數分類的損失函數選擇進行了深入分析,表明將前列腺癌分級視為回歸任務具有優越性。 5. 我們通過引入一種創新方法來整合分層Transformer中的注意力分數(平衡與任務無關和與任務相關的貢獻),增強了模型的可解釋性。
Abatract
摘要
Practical deployment of Vision Transformers in computational pathology has largely been constrained by thesheer size of whole-slide images. Transformers faced a similar limitation when applied to long documents, andHierarchical Transformers were introduced to circumvent it. This work explores the capabilities of HierarchicalVision Transformers for prostate cancer grading in WSIs and presents a novel technique to combine attentionscores smartly across hierarchical transformers. Our best-performing model matches state-of-the-art algorithmswith a 0.916 quadratic kappa on the Prostate cANcer graDe Assessment (PANDA) test set. It exhibits superiorgeneralization capacities when evaluated in more diverse clinical settings, achieving a quadratic kappa of0.877, outperforming existing solutions. These results demonstrate our approach’s robustness and practicaapplicability, paving the way for its broader adoption in computational pathology and possibly other medicalimaging tasks.
在計算病理學中,視覺Transformer(Vision Transformers)的實際部署在很大程度上受到全切片圖像(whole-slide images)龐大尺寸的限制。Transformer在處理長文檔時也面臨類似的局限,而分層Transformer(Hierarchical Transformers)的引入正是為了規避這一問題。 ? 本研究探索了分層視覺Transformer(Hierarchical Vision Transformers)在全切片圖像(WSIs)前列腺癌分級中的性能,并提出了一種跨分層Transformer智能融合注意力分數的新技術。我們性能最佳的模型在前列腺癌分級評估(PANDA)測試集上達到了0.916的加權kappa系數,與最先進算法持平。在更多樣化的臨床場景中評估時,該模型展現出更優的泛化能力,加權kappa系數達0.877,優于現有解決方案。這些結果證明了我們方法的穩健性和實際適用性,為其在計算病理學及其他醫學影像任務中的更廣泛應用奠定了基礎。
Background
背景
Method
方法
3.1. Hierarchical vision transformer
The inherent hierarchical structure within whole-slide images spansacross various scales, from tiny cell-centric regions – (16,16) pixels at0.50 microns per pixels (mpp) – containing fine-grained information, allthe way up to the entire slide, which exhibits the overall intra-tumoralheterogeneity of the tissue microenvironment. Along this spectrum,(256,256) patches depict cell-to-cell interactions, while larger regions– (1024,1024) to (4096,4096) pixels – capture macro-scale interactionsbetween clusters of cells.
3.1.?分層視覺Transformer ? 全切片圖像中固有的層級結構跨越多種尺度:從小型細胞中心區域——即0.50微米/像素(mpp)分辨率下的(16,16)像素區域,包含細粒度信息;到整個切片,呈現腫瘤內組織微環境的整體異質性。在這一尺度范圍內,(256,256)像素的補丁可描述細胞間的相互作用,而更大的區域——(1024,1024)至(4096,4096)像素——則能捕捉細胞集群間的宏觀相互作用。
Conclusion
結論
In summary, our study demonstrates the transformative potential ofHierarchical Vision Transformers in predicting prostate cancer gradesfrom biopsies. By leveraging the inherent hierarchical structure ofWSIs, H-ViTs efficiently capture context-aware representations, addressing several shortcomings of conventional patch-based methods.Our findings set new benchmarks in prostate cancer grading. Ourmodel outperforms existing solutions when tested on a dataset thatuniquely represents the full diversity of cases seen in clinical practice,effectively narrowing the generalization gap in prostate biopsy grading.This work provides new insights that deepen our understanding of themechanisms underlying H-ViTs’ effectiveness. Our results showcase therobustness and adaptability of this method in a new setting, pavingthe way for its broader adoption in computational pathology andpotentially other medical imaging tasks.
總之,本研究證實了分層視覺Transformer(Hierarchical Vision Transformers)在通過活檢樣本預測前列腺癌分級方面的變革性潛力。借助全切片圖像(WSIs)固有的層級結構,H-ViTs能夠高效捕捉具備上下文感知的表征,從而解決了傳統基于補丁的方法存在的諸多缺陷。 我們的研究結果為前列腺癌分級設立了新的基準。在測試數據集(該數據集獨特地涵蓋了臨床實踐中所見的各種病例)上,我們的模型性能優于現有解決方案,有效縮小了前列腺活檢分級中的泛化差距。 這項工作提供了新的見解,加深了我們對H-ViTs有效性背后機制的理解。研究結果展示了該方法在新場景中的穩健性和適應性,為其在計算病理學及可能的其他醫學影像任務中的更廣泛應用鋪平了道路。
Results
結果
6.1. Self-supervised pretraining
Pretraining the patch-level Transformer for 50 epochs took 3 dayson 4 GeForce RTX 3080 Ti. Fig. 4 shows the area under the curve(AUC) for teacher and student networks on the downstream patch-levelclassification dataset over pretraining epochs. Results are averagedacross the 5 cross validation folds. Early stopping was not triggeredfor any of the 5 folds. Pretraining the region-level Transformer on(4096, 4096) regions on 1 GeForce RTX 3080 Ti. More details aboutcomputational characteristics can be found in Appendix J.Classification results with CE loss are summarized in Table 2 (GlobalH-ViT) and Table 3 (Local H-ViT). Additional results for other lossfunctions can be found in Appendix A (Global H-ViT) and AppendixB (Local H-ViT). Overall, across all region sizes, models pretrainedon the PANDA dataset consistently achieve higher macro-averagedperformance than those pretrained on TCGA. Performance gains aremore significant for Global H-ViT (+68% on average) than for LocalH-ViT (+14% on average). This roots back to a common limitationof patch-based MIL: the disconnection between feature extraction andfeature aggregation. There is no guarantee the features extracted during pretraining are relevant for the downstream classification task.Allowing gradients to flow through the region-level Transformer partlyovercomes this limitation: Local H-ViT has more amplitude than GlobalH-ViT to refine the TCGA-pretrained features so that they better fit theclassification task at hand.
6.1. 自監督預訓練 ? 在4塊GeForce RTX 3080 Ti顯卡上,對補丁級Transformer進行50個 epoch的預訓練耗時3天。圖4展示了在預訓練過程中,教師網絡和學生網絡在下游補丁級分類數據集上的曲線下面積(AUC)變化。結果為5折交叉驗證的平均值,早期停止機制在5個折中均未觸發。在1塊GeForce RTX 3080 Ti顯卡上對(4096, 4096)區域的區域級Transformer進行預訓練的計算特性詳情見附錄J。 ? 采用交叉熵(CE)損失的分類結果匯總于表2(Global H-ViT)和表3(Local H-ViT)。其他損失函數的補充結果見附錄A(Global H-ViT)和附錄B(Local H-ViT)。總體而言,在所有區域大小下,基于PANDA數據集預訓練的模型在宏觀平均性能上均優于基于TCGA數據集預訓練的模型。Global H-ViT的性能提升更為顯著(平均+68%),而Local H-ViT的提升相對溫和(平均+14%)。這源于基于補丁的多實例學習(MIL)的一個常見局限:特征提取與特征聚合之間的脫節——無法保證預訓練階段提取的特征與下游分類任務相關。 ? 允許梯度通過區域級Transformer傳播可部分克服這一局限:與Global H-ViT相比,Local H-ViT擁有更大的調整空間,能夠優化TCGA預訓練特征,使其更適配當前的分類任務。
Figure
圖
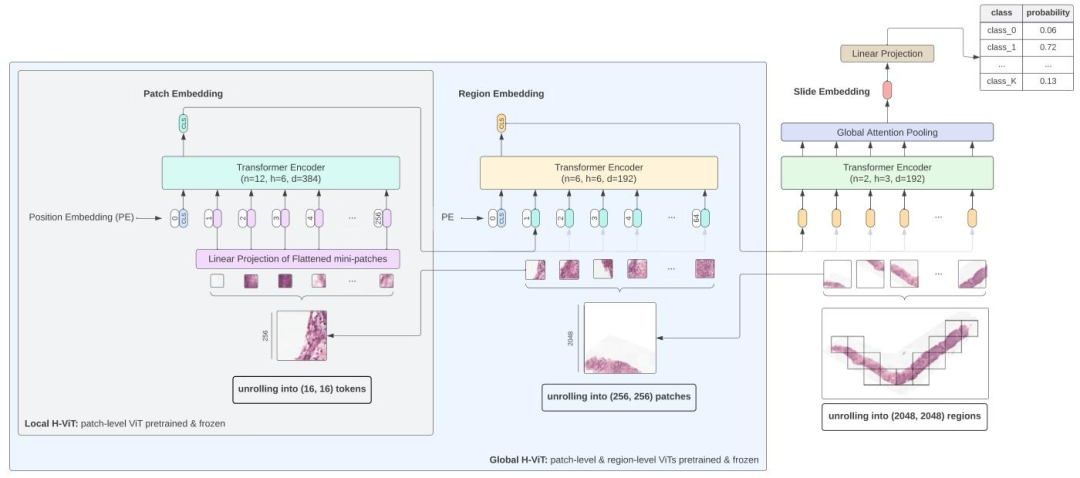
Fig. 1. Overview of our Hierarchical Vision?Transformer for whole-slide image analysis. The model processes whole-slide images at multiple scales. Slides are unrolled into nonoverlapping 2048 × 2048 regions, which are further divided into 256 × 256 patches following a regular grid. First, a pretrained ViT-S/16 (referred to as the patch-level Transformer)embeds these patches into feature vectors. These patch-level features are then input to a second Transformer (referred to as the region-level Transformer), which aggregates theminto region-level embeddings. Finally, a third Transformer (referred to as the slide-level Transformer) pools the region-level embeddings into a slide-level representation, which isprojected to class logits for downstream task prediction. We experiment with two model variants: in Global H-ViT, both the patch-level and region-level Transformers are pretrainedand frozen, with only the slide-level Transformer undergoing weakly-supervised training; in Local H-ViT, only the patch-level Transformer is frozen, while both the region-leveland slide-level Transformers are trained using weak supervision
圖1. 用于全切片圖像分析的分層視覺Transformer概述 ? 該模型以多尺度處理全切片圖像:首先將切片展開為不重疊的2048×2048區域,這些區域再按規則網格進一步劃分為256×256的補丁。第一步,預訓練的ViT-S/16(稱為補丁級Transformer)將這些補丁嵌入為特征向量;隨后,這些補丁級特征被輸入到第二個Transformer(稱為區域級Transformer),聚合為區域級嵌入;最后,第三個Transformer(稱為切片級Transformer)將區域級嵌入聚合為切片級表征,并映射為類別對數概率以用于下游任務預測。 ? 我們對兩種模型變體進行了實驗: ? - 在Global H-ViT中,補丁級和區域級Transformer均經過預訓練并固定參數,僅切片級Transformer進行弱監督訓練; ? - 在Local H-ViT中,僅補丁級Transformer固定參數,區域級和切片級Transformer均通過弱監督進行訓練。
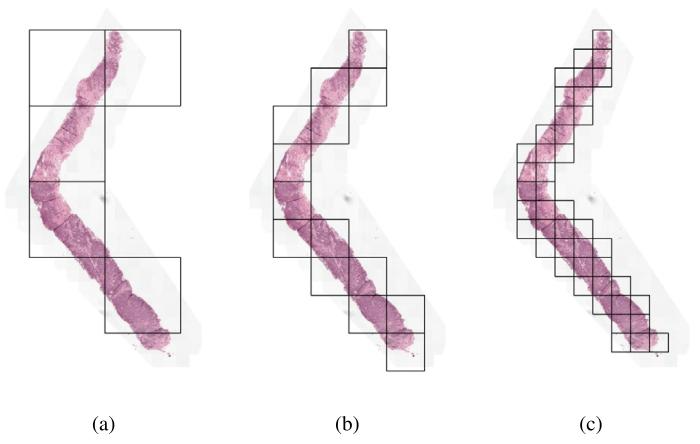
Fig. 2. Visualization of region extraction at 0.50 mpp for varying region sizes: (a)(4096, 4096) regions, (b) (2048, 2048) regions and (c) (1024, 1024) regions.
圖 2. 0.50 微米 / 像素分辨率下不同區域大小的提取可視化(a) 4096×4096 像素區域(b) 2048×2048 像素區域(c) 1024×1024 像素區域
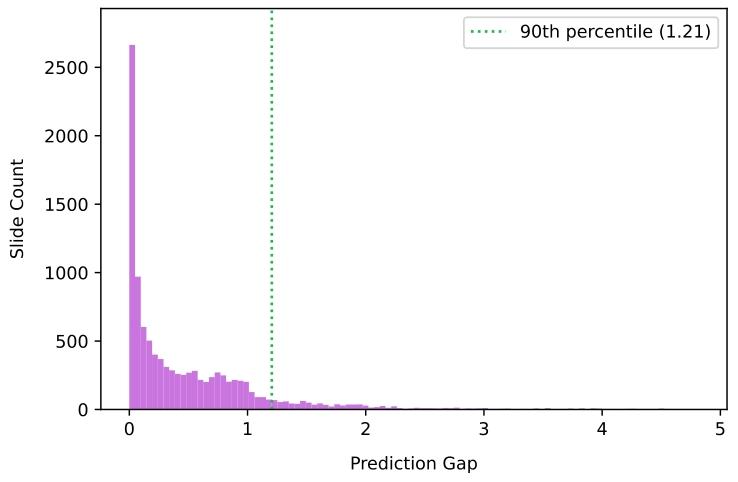
Fig. 3. Distribution of the prediction gap for PANDA development set
圖3. PANDA開發集的預測差距分布
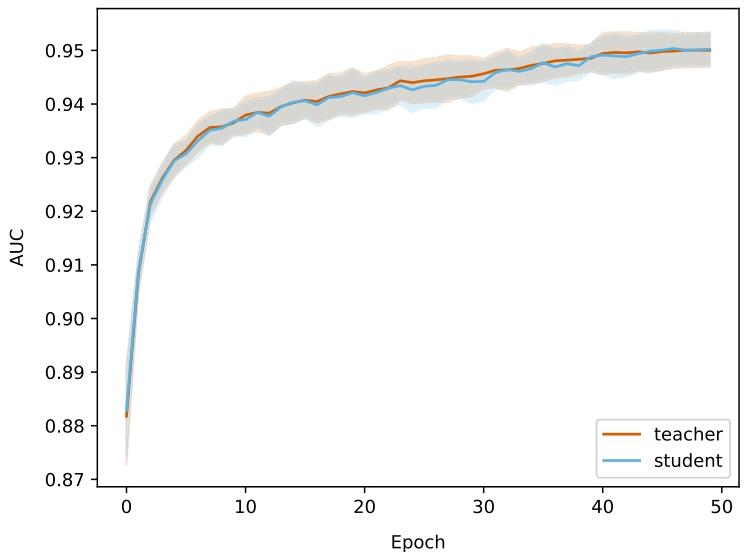
Fig. 4. Classification performance for teacher and student networks on the binaryclassification of prostate patches, used as downstream evaluation during pretraining.Lines represent the mean AUC across the 5 cross validation folds, with shaded areasindicating standard deviation.
4. 教師網絡和學生網絡在前列腺補丁二元分類任務上的分類性能(用于預訓練期間的下游評估)線條代表 5 折交叉驗證的平均 AUC(曲線下面積),陰影區域表示標準差。
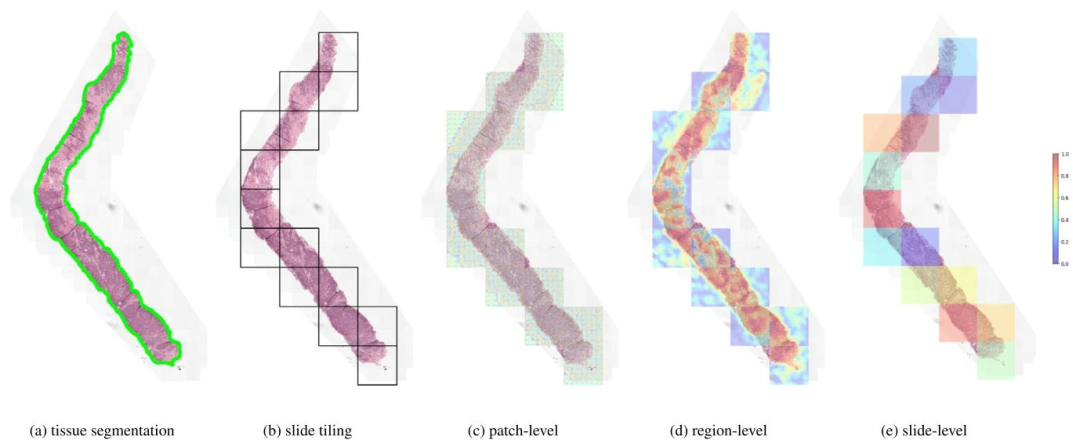
Fig. 5. Visualization of stitched attention heatmaps for each Transformer. We show the result of tissue segmentation (a) where tissue is delineated in green, and the result of slidetiling into non-overlapping (2048, 2048) regions as 0.50 mpp, keeping only regions with 10% tissue or more. For each of the three Transformers, we overlay the correspondingattention scores assigned to each element of the input sequence: (16, 16) tokens for the patch-level Transformer, (256, 256) patches for the region-level Transformer, and (2048,regions for the slide-level Transformer.
圖 5. 各 Transformer 的拼接注意力熱圖可視化我們展示了組織分割結果 (a)(綠色勾勒出組織區域),以及將切片劃分為 0.50 微米 / 像素分辨率下非重疊的 2048×2048 區域的結果(僅保留組織占比≥10% 的區域)。對于三個 Transformer,我們分別疊加了為輸入序列各元素分配的注意力分數:補丁級 Transformer 對應(16,16)像素令牌,區域級 Transformer 對應(256,256)像素補丁,切片級 Transformer 對應(2048,2048)像素區域。
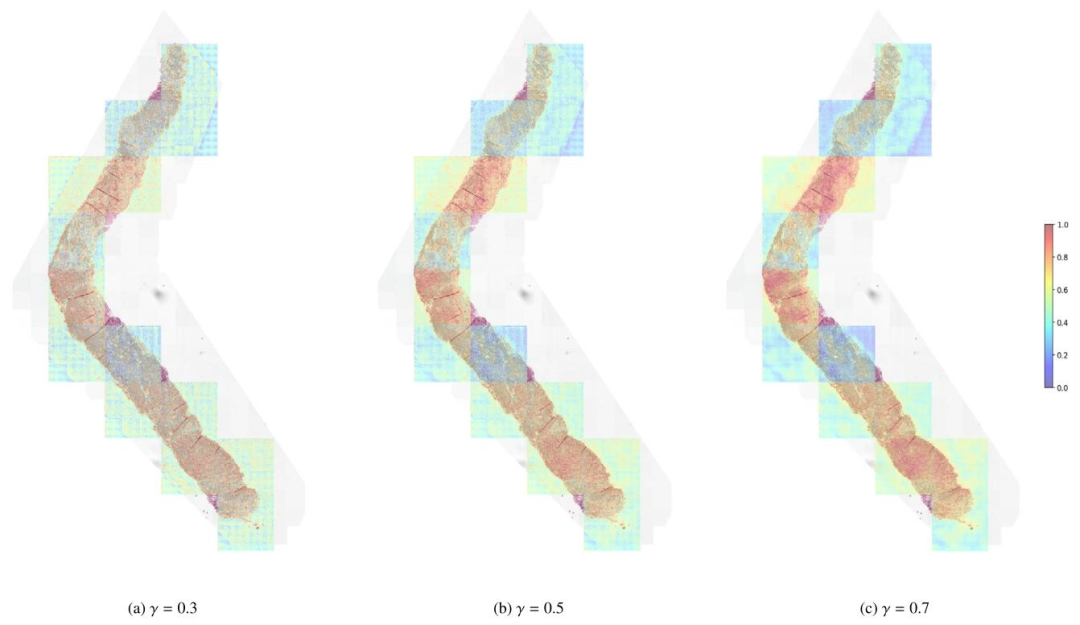
Fig. 6. Refined (2048,2048) factorized attention heatmaps of Local H-ViT for varying values of parameter 𝛾. In the context of prostate cancer grading, the relevant signal isfound within the tissue architecture, spanning intermediate to large scales. Since the patch-level Transformer primarily captures cell-level features, we recommend using 𝛾 > 0.5to emphasize coarser, task-specific features, rather than finer, task-agnostic details.
圖 6. 不同參數*𝛾*值下 Local H-ViT 的精細化(2048,2048)因子注意力熱圖在前列腺癌分級場景中,相關信號存在于組織結構中,涵蓋中等至較大尺度。由于補丁級 Transformer 主要捕捉細胞級特征,因此我們建議使用𝛾>0.5,以強調更粗略的、與任務相關的特征,而非更精細的、與任務無關的細節。
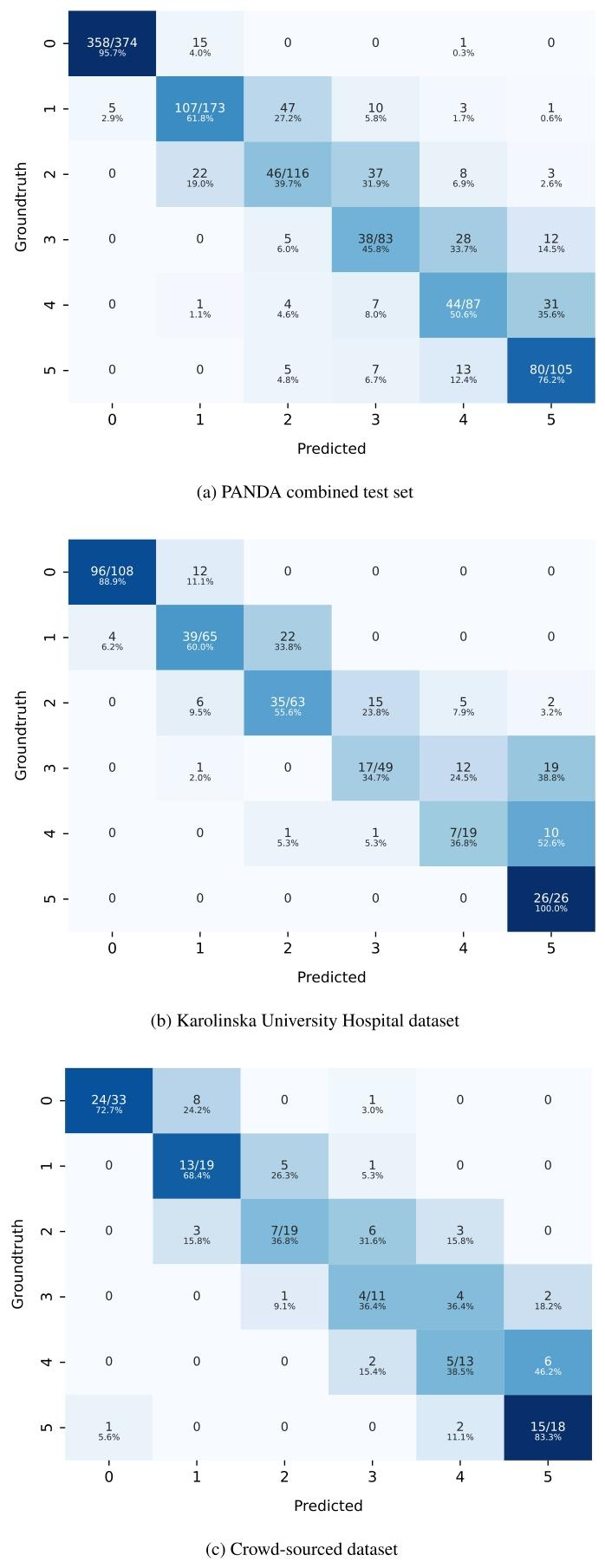
Fig. C.1. Confusion matrices of our best performing ensemble model on the 3 evaluation datasets.
圖 C.1. 最佳集成模型在 3 個評估數據集上的混淆矩陣
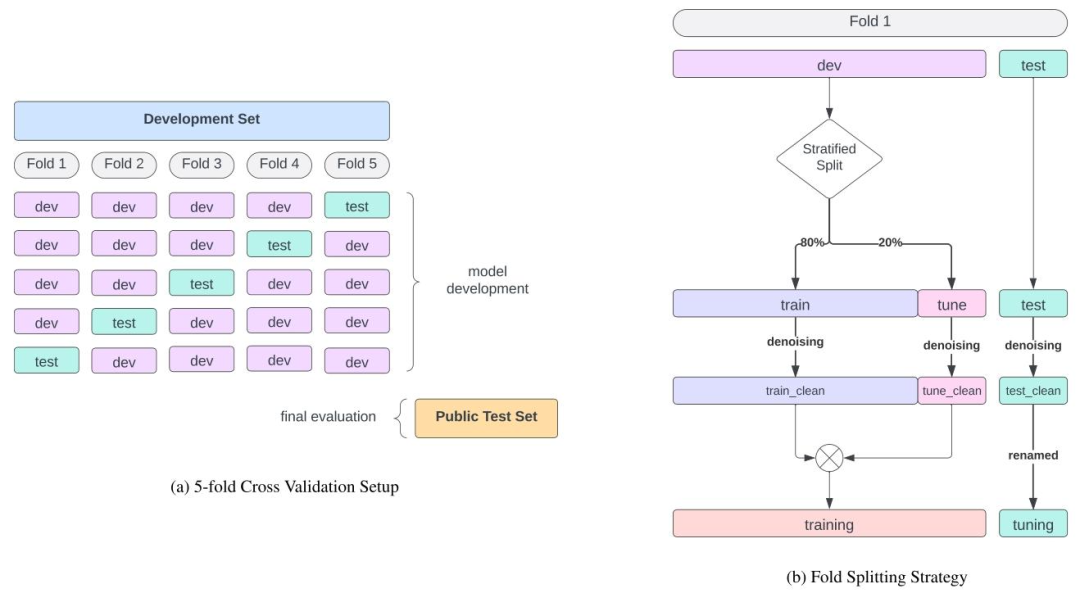
Fig. G.2. Overview of the 5-fold Cross Validation Splitting Strategy.
圖 G.2. 5 折交叉驗證分割策略概述
Fig. H.3. Visualization of four artificial blocks. (a), (c) and (d) show blocks with 2 slides. (b) shows a block with 3 slides.
圖 H.3. 四個人工區塊的可視化(a)、(c) 和 (d) 展示包含 2 張切片的區塊,(b) 展示包含 3 張切片的區塊。
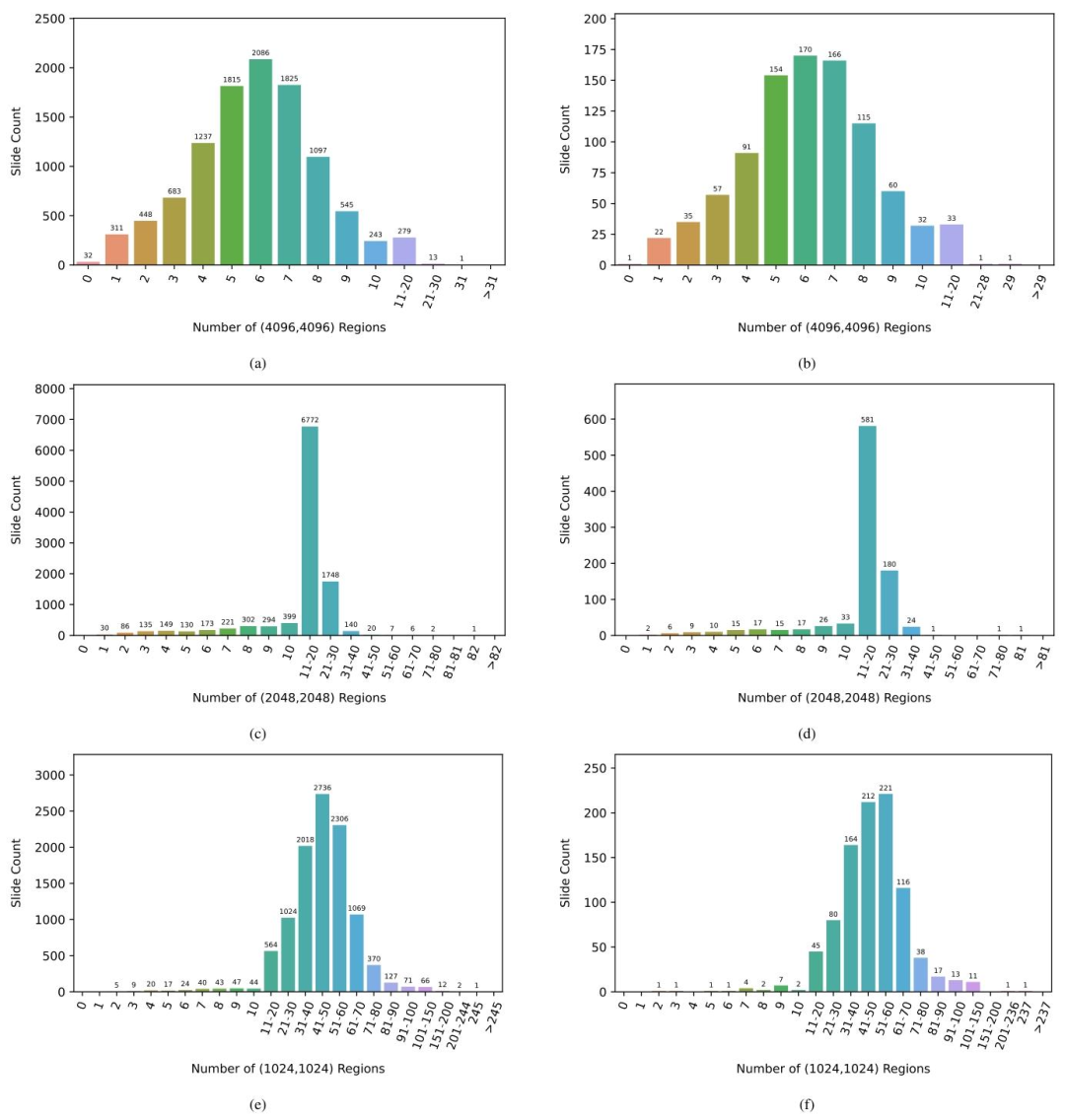
Fig. I.4. Distribution of the number of extracted regions for PANDA development set with (a) (4096, 4096) regions (c) (2048, 2048) regions (e) (1024, 1024) regions and forPANDA public test set with (b) (4096, 4096) regions (d) (2048, 2048) regions (f) (1024, 1024) regions.
圖 I.4. PANDA 開發集和公開測試集中提取的區域數量分布開發集:(a) 4096×4096 像素區域的數量分布;(c) 2048×2048 像素區域的數量分布;(e) 1024×1024 像素區域的數量分布。公開測試集:(b) 4096×4096 像素區域的數量分布;(d) 2048×2048 像素區域的數量分布;(f) 1024×1024 像素區域的數量分布。
Table
表

Table 1Scanner details, PANDA development set.
表 1 PANDA 開發集的掃描儀詳情
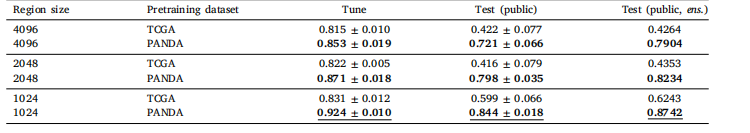
Table 2Classification performance of Global H-ViT for different pretraining configurations, obtained with cross-entropy loss. We report the mean andstandard deviation of the quadratic weighted kappa across the 5 cross-validation folds, along with the kappa score achieved by ensemblingpredictions from each fold.
表 2 不同預訓練配置下 Global H-ViT 的分類性能(采用交叉熵損失)我們報告了 5 折交叉驗證中加權 kappa 系數的均值和標準差,以及通過集成各折預測結果得到的 kappa 分數。
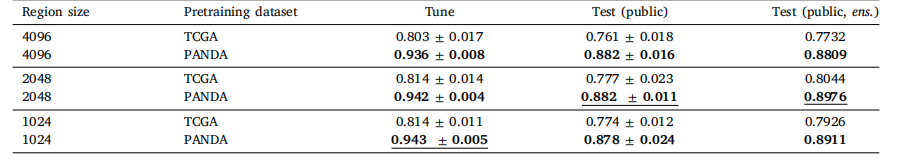
Table 3Classification performance of Local H-ViT for different pretraining configurations, obtained with cross-entropy loss. We report the mean andstandard deviation of the quadratic weighted kappa across the 5 cross-validation folds, along with the kappa score achieved by ensemblingpredictions from each fold.
表 3 不同預訓練配置下 Local H-ViT 的分類性能(采用交叉熵損失)我們報告了 5 折交叉驗證中加權 kappa 系數的均值和標準差,以及通過集成各折預測結果得到的 kappa 分數。
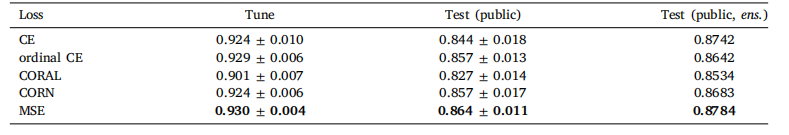
Table 4Classification performance of Global H-ViT for different loss functions, obtained with (1024, 1024) regions at 0.50 mpp. Wereport the mean and standard deviation of the quadratic weighted kappa across the 5 cross-validation folds, along with thekappa score achieved by ensembling predictions from each fold
表 4 不同損失函數下 Global H-ViT 的分類性能(采用 0.50 微米 / 像素分辨率的 1024×1024 區域)我們報告了 5 折交叉驗證中加權 kappa 系數的均值和標準差,以及通過集成各折預測結果得到的 kappa 分數
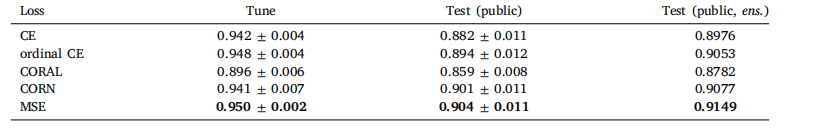
Table 5Classification performance of Local H-ViT for different loss functions, obtained with (2048, 2048) regions at 0.50 mpp. Wereport the mean and standard deviation of the quadratic weighted kappa across the 5 cross-validation folds, along with thekappa score achieved by ensembling predictions from each fold
表 5 不同損失函數下 Local H-ViT 的分類性能(采用 0.50 微米 / 像素分辨率的 2048×2048 區域)我們報告了 5 折交叉驗證中加權 kappa 系數的均值和標準差,以及通過集成各折預測結果得到的 kappa 分數。
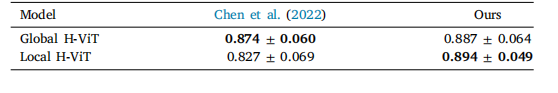
Table 6TCGA BRCA subtyping results. Both models are trained with (4096,4096) regions onthe splits from Chen et al. (2022). We report the mean and standard deviation of theAUC across the 10 cross-validation folds.
表 6 TCGA BRCA 亞型分類結果兩種模型均使用(4096,4096)區域在 Chen 等人(2022)的數據集劃分上進行訓練。我們報告了 10 折交叉驗證的 AUC(曲線下面積)均值和標準差。

Table 7Classification performance of Local H-ViT for different feature encoders, obtained with MSE loss and (2048, 2048) regions at0.50 mpp. We report the mean and standard deviation of the quadratic weighted kappa across the 5 cross-validation folds,along with the kappa score achieved by ensembling predictions from each fold.
表 7 不同特征編碼器下 Local H-ViT 的分類性能(采用 MSE 損失和 0.50 微米 / 像素分辨率的 2048×2048 區域)我們報告了 5 折交叉驗證中加權 kappa 系數的均值和標準差,以及通過集成各折預測結果得到的 kappa 分數。
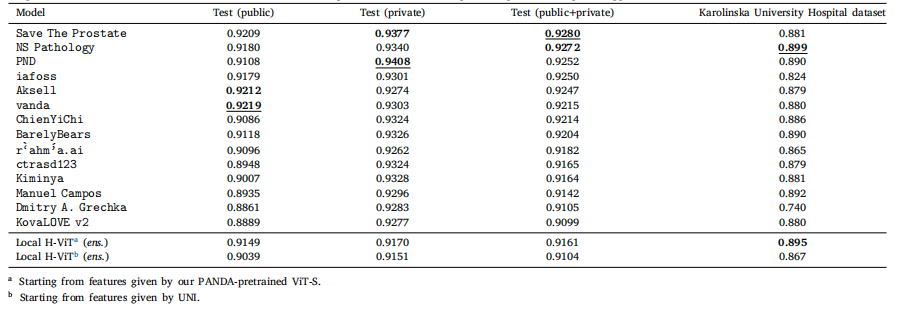
Table 8Classification performance of our best ensemble Local H-ViT models against that of PANDA consortium teams on PANDA public and private test sets, as well as Karolinska UniversityHospital dataset, used as external validation data after the challenge ended. All values are given as quadratic weighted kappa
表 8 最佳集成 Local H-ViT 模型與 PANDA 聯盟團隊模型的分類性能對比對比基于 PANDA 公開測試集、私有測試集以及卡羅林斯卡大學醫院數據集(挑戰結束后用作外部驗證數據)。所有數值均以加權 kappa 系數表示。
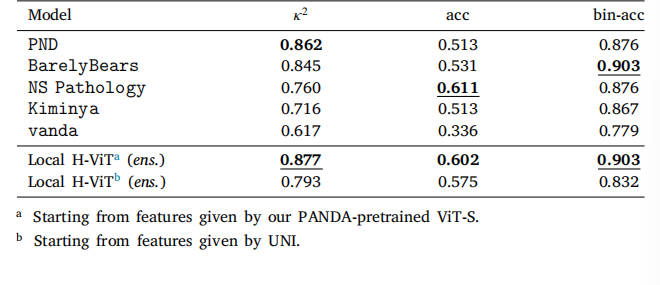
Table 9Classification performance of our ensemble Local H-ViT models compared to fivePANDA consortium teams on the crowdsourced dataset. We report quadratic weightedkappa (𝜅 2 ), overall accuracy (acc), and binary accuracy (bin-acc) for distinguishingbetween low-risk (ISUP ≤ 1) and higher-risk cases.
表 9 集成 Local H-ViT 模型與五個 PANDA 聯盟團隊模型在眾包數據集上的分類性能對比我們報告了加權 kappa 系數(𝜅2)、總體準確率(acc)以及區分低風險(ISUP ≤ 1)與高風險病例的二元準確率(bin-acc)。
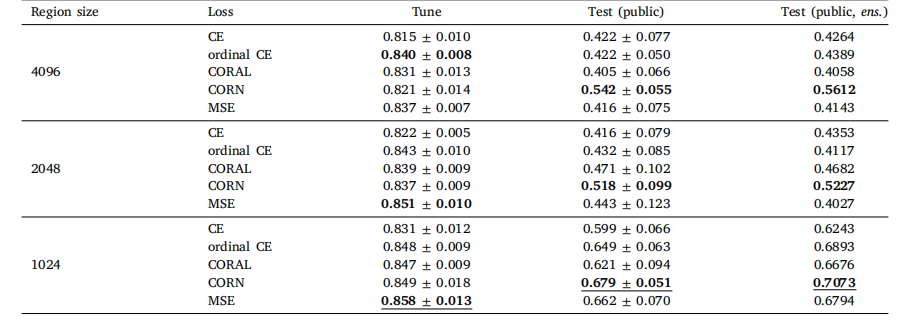
Table A.1Global H-ViT results when pretrained on TCGA dataset, for different region sizes and loss functions. We report the mean and standard deviationof the quadratic weighted kappa across the 5 cross-validation folds, along with the kappa score achieved by ensembling predictions from eachfold
表 A.1 基于 TCGA 數據集預訓練的 Global H-ViT 在不同區域大小和損失函數下的結果我們報告了 5 折交叉驗證中加權 kappa 系數的均值和標準差,以及通過集成各折預測結果得到的 kappa 分數
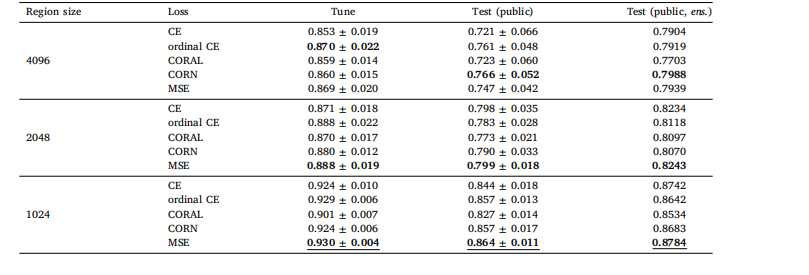
Table A.2Global H-ViT results when pretrained on PANDA dataset, for different region sizes and loss functions. We report the mean and standard deviationof the quadratic weighted kappa across the 5 cross-validation folds, along with the kappa score achieved by ensembling predictions from eachfold.
表 A.2 基于 PANDA 數據集預訓練的 Global H-ViT 在不同區域大小和損失函數下的結果我們報告了 5 折交叉驗證中加權 kappa 系數的均值和標準差,以及通過集成各折預測結果得到的 kappa 分數。
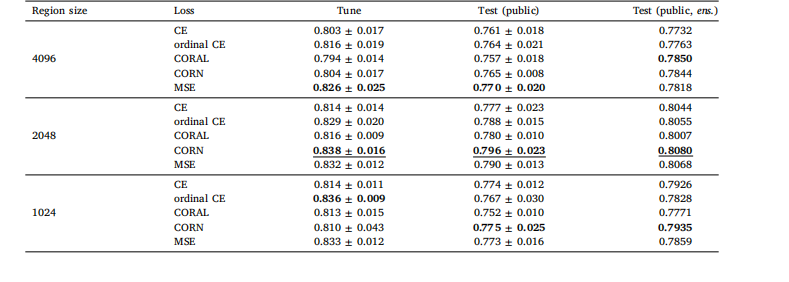
Table B.3Local H-ViT results when pretrained on TCGA dataset, for different region sizes and loss functions. We report the mean and standard deviationof the quadratic weighted kappa across the 5 cross-validation folds, along with the kappa score achieved by ensembling predictions from eachfold.
表 B.3 基于 TCGA 數據集預訓練的 Local H-ViT 在不同區域大小和損失函數下的結果我們報告了 5 折交叉驗證中加權 kappa 系數的均值和標準差,以及通過集成各折預測結果得到的 kappa 分數。
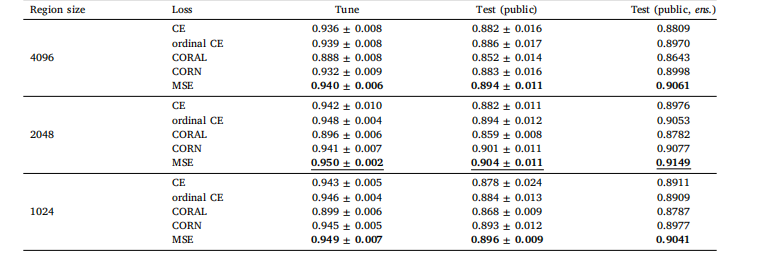
Table B.4Local H-ViT results when pretrained on PANDA dataset, for different region sizes and loss functions. We report the mean and standard deviationof the quadratic weighted kappa across the 5 cross-validation folds, along with the kappa score achieved by ensembling predictions from eachfold.
表 B.4 基于 PANDA 數據集預訓練的 Local H-ViT 在不同區域大小和損失函數下的結果我們報告了 5 折交叉驗證中加權 kappa 系數的均值和標準差,以及通過集成各折預測結果得到的 kappa 分數。
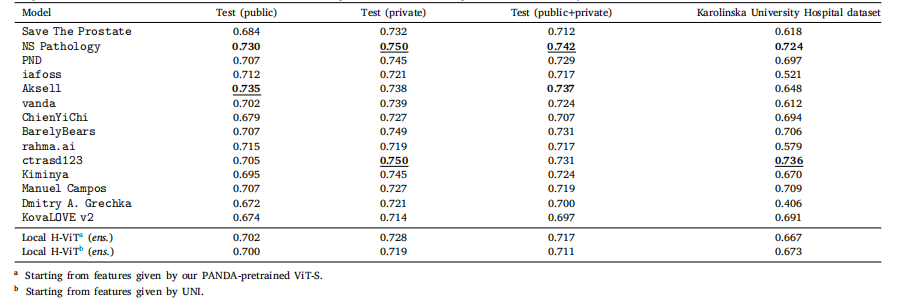
Table D.5Classification performance of our best ensemble Local H-ViT models against that of PANDA consortium teams on PANDA public and private test sets, as well as Karolinska UniversityHospital dataset, used as external validation data after the challenge ended. All values are given as overall accuracy
表 D.5 最佳集成 Local H-ViT 模型與 PANDA 聯盟團隊模型的分類性能對比對比基于 PANDA 公開測試集、私有測試集以及卡羅林斯卡大學醫院數據集(挑戰結束后用作外部驗證數據)。所有數值均以整體準確率表示。
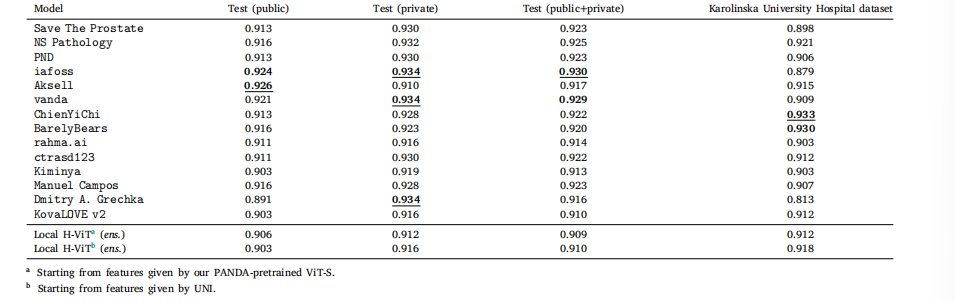
Table D.6Classification performance of our best ensemble Local H-ViT models against that of PANDA consortium teams on PANDA public and private test sets, as well as Karolinska UniversityHospital dataset, used as external validation data after the challenge ended. All values are given as binary accuracy for distinguishing between low-risk (ISUP ≤ 1) and higher-riskcases.
表 D.6 最佳集成 Local H-ViT 模型與 PANDA 聯盟團隊模型的分類性能對比對比基于 PANDA 公開測試集、私有測試集以及卡羅林斯卡大學醫院數據集(挑戰結束后用作外部驗證數據)。所有數值均以區分低風險(ISUP ≤ 1)與高風險病例的二元準確率表示。

Table E.7Average number of regions per slide, PANDA dataset
表 E.7 PANDA 數據集中每張切片的平均區域數量
Table F.8Test datasets details.
表 F.8 測試數據集詳情

Table G.9Number of slide per partition after label denoising, 5-fold CV splits.
表 G.9 標簽去噪后各劃分中的切片數量(5 折交叉驗證分割)
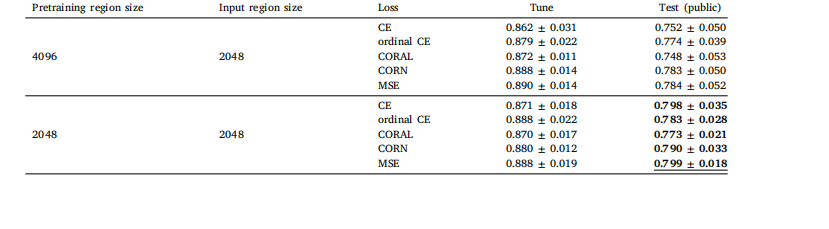
Table J.10Global H-ViT results when pretrained on PANDA dataset with (2048, 2048) regions, for different loss functions. We report quadratic weightedkappa averaged over the 5 cross-validation folds (mean ± std)
表 J.10 基于 PANDA 數據集預訓練的 Global H-ViT 在不同損失函數下的結果(采用 2048×2048 區域)我們報告了 5 折交叉驗證中加權 kappa 系數的平均值(± 標準差)。
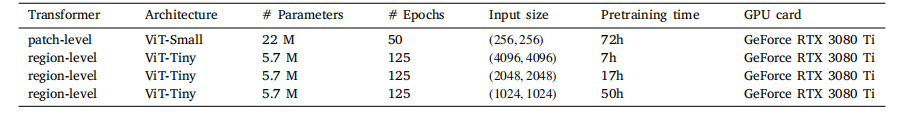
Table L.11Computational characteristics of DINO pretraining of the path-level and region-level Transformer on PANDA.
表 L.11 PANDA 數據集上路徑級和區域級 Transformer 的 DINO 預訓練計算特征

Table L.12Comparison of number of trainable parameters, training time, and GPU memory usage for Global H-ViT and Local H-ViT.
表 L.12 Global H-ViT 與 Local H-ViT 的可訓練參數數量、訓練時間及 GPU 內存使用量對比
)







)



)
的編程實現方法:1、數據包格式定義結構體2、使用隊列進行數據接收、校驗解包)

)



——標準庫函數大全(持續更新))